Isolation of Soil Microorganisms Using iChip Technology
In This Article
Summary
The iChip technique makes use of an inexpensive and simple in-situ isolation device that increases the rates of novel microorganism discovery from the soil. Novel microorganisms can be used for further study pertaining to the soil microbiome or natural product discovery, amongst other applications.
Abstract
The iChip isolation technique uses an in-situ isolation device that increases the cultivability of previously unculturable microorganisms. Microorganisms are an important source of novel chemistries and potentially bioactive molecules. However, only 1% of environmental microorganisms can be cultured using conventional laboratory methods. With the rise in antimicrobial resistance, finding new drugs to combat infections and diseases is of foremost importance, and a critical method to finding new molecules is the discovery of new microorganisms. By incubating colonies of soil microorganisms in the wells of a 96-well plate, sealed with a semipermeable membrane and incubated on top of soil, the microbes are in contact with water and growth factors from the soil, allowing for the isolation of novel microbes in a laboratory setting. After a period of domestication in an iChip, microorganisms can potentially be subcultured onto conventional media and used for further study. This device is valuable to bioactive molecule discovery and soil microbiome research and has been used previously in both applications.
Introduction
Environmental bacteria are a rich source of natural products (NPs)1. These metabolites are not vital to the survival of the microorganisms but instead are produced to facilitate their colonization by outcompeting other microorganisms in their surroundings2. Evolution has fine-tuned the chemical structures and activities of NPs, making them effective antimicrobial agents. Such an example is daptomycin, an antibiotic approved by the FDA in 20033. In recent years, microbial resistance has increased in occurrence and severity, and novel drugs like daptomycin stand as last-line treatments against infections that have developed resistance to older antibiotics4. The development of new antibiotics and other drugs is essential to keeping common infections and diseases treatable.
Although NPs often make good drug leads, the pharmaceutical industry turned to synthetic methods for drug discovery after the golden age of antibiotics in the 1950s to 1960s5. By the 1970s, the same microorganisms and antimicrobial metabolites continued to be reisolated repeatedly, with fewer and fewer novel drug candidates being discovered1,6. The soil microbiome contains great microbial diversity, but only a small number of microorganisms can be isolated under conventional laboratory conditions. The vast majority of soil microorganisms of microorganisms detected by genomics are not observed when cultured using conventional culturing methods, leading to the issue being coined "the great plate count anomaly"7. These unculturable microbes have been called microbial dark matter as they are known to exist but cannot be studied in vitro until they can be isolated as pure cultures. These unculturable microorganisms can likely produce a wealth of drug candidates, which is of great importance in the age of microbial resistance.
The iChip technique is one method that can be used to increase the recovery of novel microorganisms from the environment8,9. This technology aids in simulating the natural environment of bacteria during incubation, thereby increasing the cultivability of microorganisms that otherwise cannot survive conventional laboratory conditions10. Modified iChips have already been developed and used to isolate microorganisms from many different sources, such as soil, sediment, marine environments, and animal intestines11,12,13,14,15,16. Perhaps the most impactful case of the use of this technology has been by NovoBiotic Pharmaceuticals, where a novel bacterium, Eleftheria terrae, was discovered17. This microorganism was found to produce Teixobactin, an antibiotic of a new class, which inhibits several resistant bacterial species pertinent to human health without detected resistance development5,18. This was an impactful discovery as Teixobactin is the first new class of antibiotics discovered in decades and is a sign that this technique is a promising route for novel drug discoveries and overcoming the great plate count anomaly19. Herein, a modified iChip based on a previous publication by Berdy et al. is presented which was optimized for ease of use and contamination prevention9.
Protocol
1. Media preparation and sterilization
- Prepare and sterilize succinate minimal salts media (SMS), which consists of 0.1 g of potato starch, 1 g of casamino acids, 0.125 g of casein digest, and 15 g of bacteriological or technical agar in 1 L of water.
- Autoclave-sterilize the media and 100 mL of molecular grade water using a 20 min, 121 °C, liquid cycle.
NOTE: Ensure all other items that will come in contact with the agar-cell mixture are purchased sterile, including pipette tips and centrifuge tubes, else, autoclave them as well.
2. Modified iChip construction
- Remove the bottoms of wells in four 96-well plates using a 5 mm agar punch tool.
- Cut 0.05 µm polycarbonate membranes into 7.6 cm x 11 cm rectangles, or equal to the dimensions of the bottom of the 96-well plate.
- Using a silicone sealant, adhere 0.05 µm polycarbonate membranes to the bottom of the 96-well plates, ensuring that the adhesive seals the wells, but does not entirely cover the openings of the wells. Let dry for at least 24 h or following the instructions on the adhesive.
NOTE: Sealant must be waterproof, non-toxic, and 100% silicone. Most aquarium sealants will work well.
3. Preparation of cell suspensions
- Label four 15 mL centrifuge tubes A-D and four 50 mL centrifuge tubes E-H and add 4.5 mL sterile water to each.
- Measure 1 g of soil into a 50 mL centrifuge tube, add 10 mL of sterile water, and vortex for 10 min.
- Let the soil suspension settle for 10 min.
- Pipette 0.5 mL of the supernatant containing cells from the soil into tube A and mix thoroughly.
- Add 0.5 mL of the cell suspension in tube A to tube B and mix thoroughly. Transfer 0.5 mL of solution B to tube C and mix thoroughly. Repeat this for all remaining centrifuge tubes, completing a series of 10-fold dilutions across the eight centrifuge tubes.
- Remove 0.5 mL from tube H to have equal volume in all tubes.
NOTE: These concentrations are optimized for local soils. When attempting this procedure for the first time, carry out a larger range of dilutions to find the appropriate sample concentration.
4. Modified iChip inoculation
- Completely submerge the iChips in 95% ethanol for at least 15 min.
- Remove the plates from the ethanol and place them on sterile paper towel. While allowing the ethanol to evaporate, turn the UV sterilizer on in the laminar flow hood for 15 min to further sterilize them.
- Pipette 360 µL of sterile SMS media into the first column of the plate to act as control wells.
- Add 45 mL of SMS cooled to 50 °C to the cell suspension in tube E and mix thoroughly to combine the cell suspension and agar.
- Pipette 360 µL of the agar-cell mixture from step 4.4 into all other wells of the 96-well plate.
NOTE: A multichannel pipette is recommended as agar solidifies quickly. It is unadvised to reheat the agar-cell mixture once mixed due to the risk of killing the microorganisms. - Once the media is set, seal the top of the plates with a PCR plate cover.
- Repeat steps 3.3 to 3.6 with tubes F-H to fill a total of four iChips with ten-fold differences in concentrations.
NOTE: Media can be microwaved for 30 s with a loosened cap when needed or kept in a 60 °C hot water bath between the plates to ensure it remains molten for each.
5. Incubation
- Place the plates in a box containing approximately one inch of the soil used in step 3.2, membrane side down and place the cover.
- Incubate the plates in the covered container, in a dark place at 25 °C.
NOTE: After one week, examine the contents of the modified iChips to ensure that contamination during the setup did not occur. Contamination is indicated by the overgrowth of a single or few microorganisms in all wells of a plate. - After 6 weeks, examine the iChips. Use the plates containing growth in less than 25% of wells for colony isolation.
6. Colony isolation
- Rinse the modified iChips 3x with sterile water to remove all soil particulates.
- Wipe the top and sides of the plate with 95% ethanol, avoiding the side with the semi-permeable membrane.
- Using a sterile blade, cut through the plate cover around one well containing a colony.
- Using a sterile streaking tool, pierce the colony and streak onto SMS agar in 100 x 15 mm plates using the four-quadrant method.
- Repeat for all other colonies in the iChips that are growing one colony per well.
NOTE: For colonies that are minuscule, the plate can be placed back onto the soil and incubated until large enough to subculture as long as the plate cover was not removed around the well. - Incubate the SMS plates at 25 °C and monitor growth.
- Examine colonies to ensure axenic cultures have been isolated.
7. Identification of microorganisms
- Sequence the genomes of the isolates recovered20.
- Evaluate the genomes using JSpeciesWS or another genome-based identification platform to determine the degree of similarity of each isolate to known microorganisms in GenomesDB.
- Categorize isolates below the community accepted thresholds for species identification as putatively new.
Results
A visual overview of the protocol is shown in Figure 1. A successful modified iChip experiment results in at least one plate containing growth in less than 25% of the wells, with no growth occurring in the control wells. This number of wells with growth generally ensures that single colonies are isolated in wells. Additionally, the number of colonies, or the number of wells containing colonies, should decrease tenfold with each subsequent plate prepared from the series of dilutions. Representative results are shown in Figure 2. where plates 7 and 8 are successful iChips as they contain growth in less than 25% of wells, and the percentage of wells containing colonies decreases with each plate, as shown in Table 1. If all plates in a trial contain more than one colony per well or no colonies in any wells, that constitutes a negative outcome.
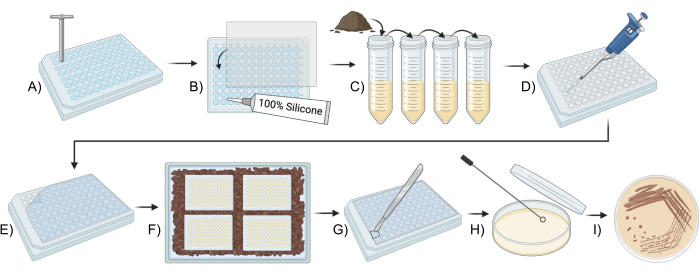
Figure 1: Schematic overview for using a modified iChip to culture soil microbes. (A,B) Construction of the plate (Step 2). (C) Preparation of inoculum (Step 3). (D,E) iChip setup (Step 4). (F) Incubation of the iChip (Step 5). (G-I) Transfer of the isolates to conventional media (Step 6). Please click here to view a larger version of this figure.
| Ichip | Concentration of soil in agar suspension | Wells with colonies | Colony-forming Units |
| E | 5.0 x 10-6 g/mL | All | Too many to count |
| F | 5.0 x 10-7 g/mL | All | Too many to count |
| G | 5.0 x 10-8 g/mL | 22 | 22 |
| H | 5.0 x 10-9 g/mL | 6 | 6 |
Table 1: Representative data from a successful iChip experiment where the number of wells containing colonies decreases with each dilution in series.
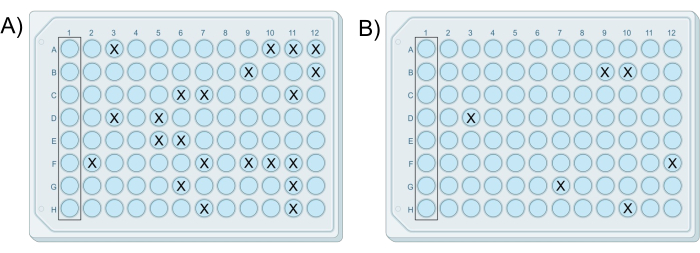
Figure 2: Exemplary results of two successful modified iChips. The iChips were from the same experiment in which (A) the plate prepared from cell-agar mixture G contains 22 colonies, and (B) the plate prepared from cell-agar mixture H contains six colonies, consistent with the 10-fold dilution difference between mixtures. Please click here to view a larger version of this figure.
One can visualize colonies with the naked eye or microscopy, as shown in Figure 3A-G. Colonies may be growing on the surface of the agar, as seen in Figure 3B or embedded in the agar, such as the colony in Figure 3F. There may be cases in which there are two colonies in the same well in a successful modified iChip, such as in Figure 3G. In some cases, the colonies are far enough apart that they can be pierced with a needle separately and subcultured; however, it is unlikely those in Figure 3G could be easily cultured as axenic microorganisms. Examples of colonies transferred from the iChip onto conventional SMS media are shown in Figure 4.
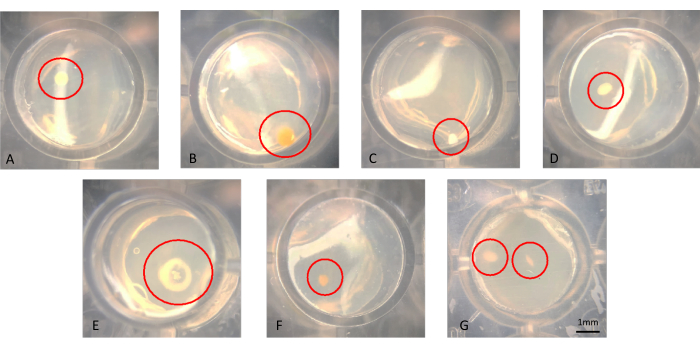
Figure 3: Representative microscopic view of colonies growing in an iChip after 6 weeks of incubation. (A-F) Single colonies are observed, which should be subcultured from, and in (G) two colonies are observed in close proximity, which are difficult to obtain as axenic cultures. Please click here to view a larger version of this figure.
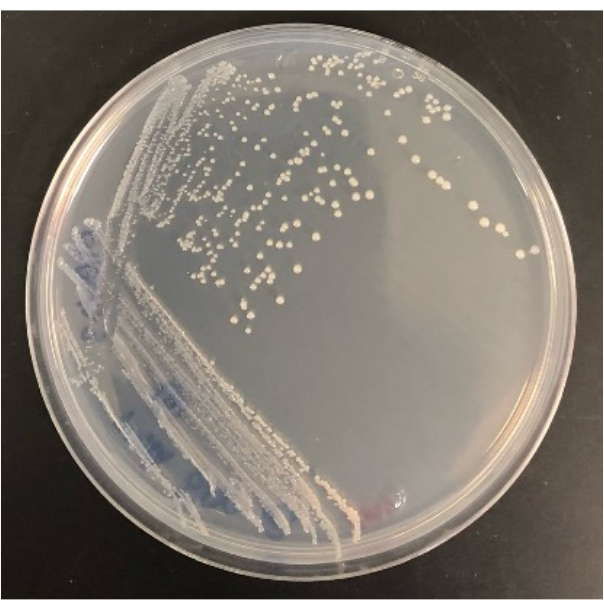
Figure 4: Representative result of a bacterium isolated from the modified iChip once growing on an SMS agar plate. Please click here to view a larger version of this figure.
The ability to isolate colonies from the modified iChip onto conventional agar plates also determines the experiment's success. Not all colonies growing in the modified iChip will survive the transfer to conventional plates. In an iChip, colonies are exposed to growth factors and soil nutrients, increasing their cultivability. For some microorganisms, a period of domestication is not enough to make them culturable in conventional conditions. The recovery rate of colonies from an iChip to plates will vary from experiment to experiment, but some should grow in plates as this method will also culture easily culturable microorganisms. For example, the modified iChips in Figure 2 contained 28 colonies in total, and the number of colonies that grew on agar plates after subculture was 16, while 12 did not survive the transfer. Isolates can be identified and used for further study once cultured on full-sized agar plates, as shown in Figure 4.
A successful modified iChip experiment should result in the isolation of microorganisms that differ from known microorganisms at the species level. The degree of similarity of each isolate to known microorganisms is found by comparing the isolated DNA to the genomes of known microorganisms. The tetra Z scores of the closest related microorganisms to each isolate describe the percent similarity. Figure 5 shows that of the twelve bacteria isolated, four were new species or higher, two were likely new strains, and six were known microorganisms. The recovery of several novel species or strains is consistent with previously published iChip experiments, indicating that this modified iChip construction does not impede the increase in the recovery of novel species11.
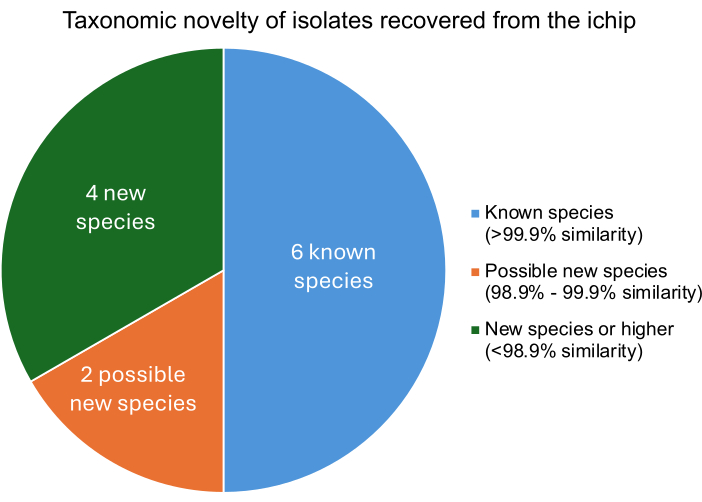
Figure 5: The taxonomic classification level of isolates recovered from an iChip experiment. Percent similarity was determined based on the tetra Z scores, which describe the isolates similarity to known bacteria where >99.9% similarity indicates a species match, 98.9%-99.9% similarity indicates a possible new species, and <98.9% similarity indicates a new species or higher. Please click here to view a larger version of this figure.
Discussion
Numerous methods, such as genome mining and examining silent biosynthetic pathways, have facilitated the discovery of novel bioactive chemicals in recent years21,22. However, NPs discovered using such methods often exhibit high structural similarity to known compounds. Gaining access to previously uncultured microorganisms will unlock greater chemical diversity and NPs with novel modes of action that can better aid in combatting microbial resistance. The iChip isolation technique has been shown to increase the cultivability of novel microorganisms, which can be used to build libraries of microorganisms for the discovery of novel NPs.
The earliest conceptualization of an iChip was published in 2002, which consisted of single metal rings containing an agar cell mixture sealed with semi-permeable membranes and incubated in situ23. It was later iterated upon in 2010 to contain a small chip with many wells11, followed by the development of an inexpensive iChip constructed from common lab materials in an influential Nature Protocols publication in 20179. Several modifications were made to the Nature protocol in this publication to improve its practicality and ease of use. The Nature protocol involves gluing a semi-permeable membrane to both sides of a chip compared to this method, which uses an adhesive plate cover on one side. Issues may arise when using silicone glue to attach the membrane after the iChip is loaded. To our knowledge, all non-toxic silicone adhesives emit acetic acid as they set, which may impact cell viability24. The protocol outlined in this video also significantly reduces the amount of handling required after inoculation by using the PCR plate cover to seal the filled plate, further ensuring sterility and reducing the set-up time. Based on the identification of multiple novel isolates from this experiment, the use of only a single semi-permeable membrane does inhibit the increase in rates of novel organism discovery that has been reported for iChips constructed with two semipermeable membranes. However, further experiments with a greater sample size would be required to quantify the impact.
Another method modification is to only subculture from the modified iChips that contain growth in less than 25% of the wells. If colonies grow in the majority of the wells of a plate, there are likely some wells that contain multiple microorganisms, even if not easily visible. During method development it was found that it was largely impossible to obtain axenic cultures when subculturing from wells containing more than one colony. The isolation of non-axenic cultures presents significant issues downstream in terms of bioactivity assays and identification. Thus, for simplicity, only modified iChips with colonies growing in a portion of the wells are recommended to be subcultured from.
The most significant issue that can arise with iChip methods is the contamination of wells or entire iChips with other microorganisms. Contamination is indicated by colonies growing in the control wells, or the same microorganism growing across several wells or an entire area of the modified iChip. The source of contamination could be due to inadequate sterilization of materials during the set-up, or improper aseptic technique. In such cases, ensure all materials used are autoclave or ethanol sterilized as indicated in the method, and ensure no contact occurs between any non-sterile items and the modified iChip other than the agar-cell mixture. If an overgrowth of a single microorganisms is observed at the bottom or top of multiple modified iChip wells it is most likely the result of an incomplete seal between the wells and membrane. In this case, ensure that the adhesive used is 100% silicone, which does not degrade in ethanol, and ensure the semipermeable membrane and PCR plate cover are completely sealed around each well during construction.
The dilution series used in the current protocol should provide adequate dilution for most soil types with a storage period of less than one week as it includes a thousand-fold range of dilutions. However, there can be significant variation in microorganism counts between soil types. If the control wells contain no growth but multiple colonies are growing in each well of all four modified iChips, the cell concentrations used were not low enough. The dilution series should be modified to reach lower cell concentrations in the cell-media mixtures used to set up the modified iChips. Similarly, if no growth is observed in any of these plates, the concentrations used in the cell-media mixtures should be increased. Alternatively, it is possible that the temperature of the media used was too high for the survival of microorganisms. In such a case, the media should be allowed to cool as much as possible without solidifying before being added to the diluted cell suspension.
The technology is an important advance toward overcoming the great plate count anomaly. However, it is still limited by the unsuitability of conventional culturing techniques, as indicated by the number of microorganisms that do not survive the transfer from the modified iChip to conventional agar plates. Previous publications have reported that multiple rounds of subculture and incubation in iChips further increase the cultivability of the microorganisms. The extended domestication time and exposure to soil growth factors while on agar in the iChip increases the chances of a colony growing on agar alone. However, this approach has not been reported to yield a higher likelihood of novel microorganisms than a single iChip incubation25,26.
Many other tactics are being explored and modified to increase the cultivability of novel microorganisms. For example, making an entire iChip device out of a semi-permeable material has been proposed to facilitate the co-culture of microorganisms in neighboring wells27. That being said, an advantage of construction outlined in this publication is its low cost with the cost of building one plate equating to approximately $12 ($4 per 96-well plate, $8 per membrane, $2 per PCR cover). Furthermore, its simple construction makes it an uncomplicated tool when used as described and provides many possibilities for customization. Though this protocol uses a medium selective for bacteria, the experimental set-up can theoretically be tuned to target a desired microbial population by modifying the medium used, such as using bacteria-suppressing media to target fungi, or low nutrient agar for sporulating microorganisms.
Disclosures
The authors have declared to have no conflicts of interest.
Acknowledgements
We gratefully acknowledge the J-base funding (J-001757, J-001842) provided through Agriculture and Agri-Food Canada, which made this project possible. We thank Brett van Heyningen for filming the video content for this protocol. We would also like to thank Ron Matters for collecting soil samples used in the experiments described in this publication.
Materials
| Name | Company | Catalog Number | Comments |
| 0.1-20 µL pipette tips | VWR | 76322-158 | Pack of 768 |
| 0.2 mL PCR tubes | ThermoFisher Scientific | AB0337 | Case of 1000 |
| 0.5-5 mL single channel pipette | VWR | CA11020-004 | |
| 1 L glass bottle | Millipore Sigma | CLS13951L | Must be autoclaveable. |
| 100 x 15 mm Petri plates | VWR | 25384-342 | Case of 500 |
| 100% Silicone sealant | Marineland | 31003 | |
| 1000 µL multichannel pipette tips | ThermoFisher Scientific | 9401113 | Case of 960 |
| 100-1000 µL pipette tips | VWR | 76322-164 | Pack of 768 |
| 100-1000 µL single channel pipette | VWR | 76169-240 | |
| 100-1200 µL multichannel pippette | ThermoFisher Scientific | 46300800 | Must have 360 µL volume capacity. |
| 1-10 µL single channel pipette | VWR | 76169-232 | |
| 1-5 mL pipette tips | VWR | CA11020-008 | Pack of 500 |
| 15 mL sterile centrifuge tubes | VWR | CA21008-918 | Case of 500 |
| 16s rRNA-F primer (AGAGTTTGATCCTGGCTCAG) – 10mM | Integrated DNA Technologies | 51-01-19-06 | 10 µg |
| 16s rRNA-R primer (ACGGCTACCTTGTTACGACTT) – 10mM | Integrated DNA Technologies | 51-01-19-07 | 10 µg |
| 4 mm cork borer | VWR | 470121-860 | |
| 50 mL sterile centrifuge tubes | VWR | CA21008-940 | Case of 500 |
| 95% ethanol | Thermo Fisher Scientific | A412-500 | 500 mL |
| 96-well plate | VWR | 10062-900 | Case of 100 |
| Autoclave | Cole-Parmer | UZ-01850-34 | 8 L, 115 VAC |
| Bacteriological agar | ThermoFisher Scientific | 443570010 | 1 kg |
| bin | Thomas Scientific | 1216H91 | 5 bins per pack |
| Casamino acids | ThermoFisher Scientific | 223120 | 500 g |
| Casein digest | ThermoFisher Scientific | 211610 | 500 g |
| Electrofluoresis grade agarose | Thermo Fisher Scientific | J66501.30 | 250 g |
| iBright FL1500 Imaging System | ThermoFisher Scientific | A44115 | |
| Laminar flow hood | CleanTech | 1000-6-A | |
| Minion Nanopore Sequencer | Oxford Nanopore Technologies | MinIon Mk1C | |
| NanoDrop One/One Microvolume UV-Vis Spectrophotometer | ThermoFisher Scientific | ND-ONE-W | |
| Nuclease Free Water | Thermo Fisher Scientific | AM9937 | 10 x 50 mL |
| Nucleobond HMW DNA kit | Takara | 740160.2 | |
| Paper towel | VWR | 89402-824 | |
| Phusion Green Hot Start II High-Fidelity PCR Master Mix | Thermo Fisher Scientific | F566L | 500 Reactions |
| Potato starch | ThermoFisher Scientific | 419690025 | 2.5 kg |
| QIAGEN CLC Genomics Workbench Software or similar | Qiagen | ||
| Rapid Barcoding 24 Kit | Oxford Nanopore Technologies | SQK-RBK114.24 | |
| SimpliAmp thermal cycler | Applied Biosystems | A24811 | |
| Sterile Inoculation loops with needle | VVWR | 76534-512 | Case of 1000 |
| Sterile surgical blade | VWR | 76457-444 | |
| SYBR Safe, or simmilar | ThermoFisher Scientific | S33101 | |
| UltraPure Agarose | ThermoFisher Scientific | 16500-500 | |
| Vortex | VWR | 76549-928 | Must accomadate 15 and 50 mL centrifuge tubes |
| VWR Stereo Zoom Trinocular Microscope | VWR | 89404-476 | |
| Whatman Nuclepore Track-Etched Membranes | Millipore Sigma | WHA113502 | L x W 8 in. x 10 in., pore size 0.03 μm |
References
- Newman, D. J., Cragg, G. M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 83 (3), 770-803 (2020).
- Fouillaud, M., Dufosse, L. Microbial secondary metabolism and biotechnology. Microorganisms. 10 (1), 123 (2022).
- Rizzetto, G., et al. Role of daptomycin in cutaneous wound healing: A narrative review. Antibiotics. 11 (7), 944 (2022).
- Miethke, M., et al. Towards the sustainable discovery and development of new antibiotics. Nat Rev Chem. 5 (10), 726-749 (2021).
- Gunjal, V., Thakare, R., Chopra, S., Reddy, D. S. Teixobactin: A paving stone toward a new class of antibiotics. J Med Chem. 63, 12171-12195 (2020).
- Atanasov, A. G., Zotchev, S. B., Dirsch, V. M., Supuran, C. T. Natural products in drug discovery: Advances and opportunities. Nat Rev Drug Discov. 20 (3), 200-216 (2021).
- Epstein, S. S. The phenomenon of microbial uncultivability. Curr Opin Microbiol. 16 (5), 636-642 (2013).
- Wright, G. An irresistible newcomer. Nature. 517, 422-444 (2015).
- Berdy, B., Spoering, A., Ling, L., Epstein, S. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat Protoc. 12 (10), 2232-2242 (2017).
- Jung, D., et al. Triggering growth via growth initiation factors in nature: A putative mechanism for in situ cultivation of previously uncultivated microorganisms. Front Microbiol. 12, 537194 (2021).
- Nichols, D., et al. Use of ichip for high-throughput in situ cultivation of "uncultivable" microbial species. Appl Environ Microbiol. 76 (8), 2445-2450 (2010).
- Megaw, J., Kelly, S. A., Thompson, T. P., Skvortsov, T., Gilmore, B. F. Profiling the microbial community of a triassic halite deposit in Northern Ireland: An environment with significant potential for biodiscovery. FEMS Microbiol Lett. 366 (22), fnz242 (2019).
- Vitorino, I., et al. Novel and conventional isolation techniques to obtain planctomycetes from marine environments. Microorganisms. 9 (10), 2078 (2021).
- Moote, P., Polo, R. O., Uwiera, R. R. E., Inglis, G. D. Comparison of strategies for isolation anaerobic bacteria from the porcine intestine. Appl Environ Microbiol. 87 (9), e00088-e00121 (2021).
- Vitorino, I. R., et al. Rhodopirellula aestuarii sp. Nov., a novel member of the genus rhodopirellula isolated from brackish sediments collected in the tagus river estuary, Portugal. Syst Appl Microbiol. 45 (6), 126360 (2022).
- Ding, H., et al. Hanstruepera marina sp. Nov. and Hanstruepera flava sp. Nov., two novel species in the family flavobacteriaceae isolated by a modified in situ cultivation technique from marine sediment. Front Microbiol. 13, 957397 (2022).
- Ling, L. L., et al. A new antibiotic kills pathogens without detectable resistance. Nature. 517 (7535), 455-459 (2015).
- Qi, Y. K., et al. Discovery, synthesis, and optimization of teixobactin, a novel antibiotic without detectable bacterial resistance. J Pept Sci. 28 (11), e3428 (2022).
- Piddock, L. J. Teixobactin, the first of a new class of antibiotics discovered by ichip technology. J of Antimicrob Chemother. 70 (10), 2679-2680 (2015).
- Gong, L., Wong, C. H., Idol, J., Ngan, C. Y., Wei, C. L. Ultra-long read sequencing for whole genomic DNA analysis. J Vis Exp. (145), e58954 (2019).
- Covington, B. C., Xu, F., Seyedsayamdost, M. R. A natural product chemist's guide to unlocking silent biosynthetic gene clusters. Annu Rev Biochem. 90, 763-788 (2021).
- Lee, N., et al. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput Mol Biol. 18, 1548-1556 (2020).
- Kaeverlein, T., Lewis, K., Epstein, S. S. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science. 296 (5570), 1127-1129 (2002).
- Trcek, J., Mira, N. P., Jarboe, L. R. Adaptation and tolerance of bacteria against acetic acid. Appl Microbiol Biotechnol. 99 (15), 6215-6229 (2015).
- Kurm, V., Van Der Putten, W. H., Hol, W. H. G. Cultivation-success of rare soil bacteria is not influenced by incubation time and growth medium. PLOS One. 14 (1), e0210073 (2019).
- Buerger, S., et al. Microbial scout hypothesis and microbial discovery. Appl Environ Microbiol. 78 (9), 3229-3233 (2012).
- Lodhi, A. F., Zhang, Y., Adil, M., Deng, Y. Antibiotic discovery: Combining isolation chip (ichip) technology and co-culture technique. Appl Microbiol Biotechnol. 102 (17), 7333-7341 (2018).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved