È necessario avere un abbonamento a JoVE per visualizzare questo. Accedi o inizia la tua prova gratuita.
RNAi Plating for C. elegans Feeding: A Technique to Induce Target dsRNA Expression in E. coli
In questo articolo
Overview
This video describes the principles behind RNAi treatment in C. elegans and demonstrates a protocol to knockdown lin-35 in a transgenic worm strain.
Protocollo
This protocol is excerpted from Kolundzic, et al, Application of RNAi and Heat-shock-induced Transcription Factor Expression to Reprogram Germ Cells to Neurons in C. elegans, J. Vis. Exp. (2018).
- Solution Preparation
- NGM-Agar plates (1 L)
- Add 3 g of NaCl, 2.5 g of peptone media (e.g., Bacto-Peptone), and 20 g of agar. After autoclaving, add 1 mL of cholesterol (5 mg/mL in 95% EtOH stock), 1 mL of 1 M MgSO4, 1 mL of 1 M CaCl2, 25 mL of 1 M K2PO4, and 1 mL of amphotericin B (2.5 mg/mL stock).
- NGM-Agar RNAi plates (1 L)
- Add 3 g of NaCl, 2.5 g of peptone media, and 20 g of agar. After autoclaving add, 1 mL of cholesterol (5 mg/mL in 95% EtOH stock), 1 mL of 1 M MgSO4, 1 mL of 1 M CaCl2, 25 mL of 1 M K2PO4, 1 mL of amphotericin B (2.5 mg/mL stock), 1 mL of 1 M IPTG, and 1 mL carbenicillin (50 mg/mL).
- LB-Agar (1 L)
- Add 10 g of peptone media, 5 g of yeast extract, 5 g of NaCl, 20 g of agar, 10 mL of 1 M Tris pH 8.0. Add H2O to 1 L. After autoclaving, add 1 mL of 50 mg/mL carbenicillin and 2.5 mL of 5 mg/mL tetracycline.
- LB Medium (1 L)
- Add 10 g of peptone media, 5 g of yeast extract, 5 g of NaCl, and 10 mL of 1 M Tris pH 8.0. Add H2O to 1 L. After autoclaving, add 1 mL of 50 mg/mL carbenicillin.
- M9-buffer (1 L)
- Add 6.0 g of Na2HPO4, 3 g of KH2PO4, 5 g of NaCl, and 50 mg of gelatin. Before usage, add 1 mL of 1 M MgSO4.
- Bleaching solution (10 mL)
- Add 1 mL of NaClO and 2 mL of 5 N NaOH. Add H2O to 10 mL.
- NGM-Agar plates (1 L)
- Preparation of RNAi Plates
NOTE: C. elegans is typically cultured in the laboratory on 6 cm Petri plates containing 7.5 mL of Nematode Growth Medium Agar (NGM-Agar). This protocol is optimized for worms kept at 15 °C. To prepare plates with NGM, use standard sterile techniques to prevent fungal and bacterial contamination. The following is the protocol to prepare NGM plates supplemented with reagents for RNAi.- Prepare 6-cm NGM-Agar RNAi plates containing 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 50 µg/mL carbenicillin. Let them dry for 24 – 48 h at room temperature in the dark12.
- Keep RNAi plates at 4 °C in the dark. Do not use if older than 14 days.
- Select the RNAi bacteria (Escherichia coli HT115) clone containing the L4440 plasmid with the lin-53 gene DNA sequence from the frozen glycerol stock from the available RNAi library on LB-agar plates containing 50 µg/mL carbenicillin and 12.5 µg/mL tetracycline by using the three-phase streaking pattern. Grow the bacteria at 37 °C overnight. This clone allows IPTG-dependent production of lin-53 dsRNA in the bacteria.
- Additionally, grow RNAi bacteria that contain the L4440 plasmid without any gene sequence as the empty vector control.
- The next day, pick a single colony from lin-53 or empty vector LB-agar plates using a 200 µL pipette tip and inoculate each of them into a separate culture tube containing 2 mL of liquid LB medium supplemented with 50 µg/mL carbenicillin. Grow cultures overnight at 37 °C until they reach an optical density (OD) at 600 nm of 0.6 – 0.8. Measure the OD using a spectrophotometer.
CAUTION: The liquid LB media should not contain tetracycline in contrast to the LB-agar plate used for streaking the bacteria from the glycerol stock, since inclusion of tetracycline during feeding leads to a decreased RNAi efficiency. - Add 500 µL of each bacterial culture (lin-53 or empty vector) to 6 cm NGM-Agar RNAi plates using a multipipette. Incubate plates with a closed lid overnight at room temperature in the dark to dry. During this time, the IPTG in the NGM plates will induce production of dsRNA in the bacteria.
- Use at least 3 plates per bacterial culture per experiment. This will provide 3 technical replicates for each experiment.
- Store dried NGM-agar RNAi plates with bacteria at 4 °C in the dark for up to two weeks.
- Prepare 6-cm NGM-Agar RNAi plates containing 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 50 µg/mL carbenicillin. Let them dry for 24 – 48 h at room temperature in the dark12.
- Preparation of C. elegans Strain BAT28
- Maintain the worm strain BAT28 containing the transgenes otIs305 [hsp-16.2prom::che-1, rol-6(su1006)] and ntIs1 [gcy-5prom::gfp] on OP50 bacteria using standard NGM-Agar plates at 15 °C.
NOTE: For detailed protocol on Nematode culture, see. The rol-6(su1006) is a dominant allele and commonly used phenotypic injection marker16 that causes the typical rolling movement. It is used in the otIs305 transgene to track hsp-16.2prom::che-1.- Keep the BAT28 strain at 15 °C at all times in order to prevent precautious activation of the hsp-16.2prom::che-1 transgene. Reduce exposure to temperatures higher than 15 °C while handling the strain as much as possible.
- To age-synchronize worms, use the bleaching technique.
- Wash off 6 cm NGM-Agar plates containing adults and eggs of BAT28 using 900 µL M9 buffer. Pellet worms by centrifuging at 900 x g for 1 min. Remove the supernatant.
- Add 0.5 – 1 mL of bleaching solution and shake the tube for approximately 1 min until adult worms start to burst open. Monitor using a standard stereomicroscope.
- Pellet worms by centrifuging at 900 x g for 1 min. Remove the supernatant.
- Wash the worm pellet 3 times by adding 800 µL of M9 buffer and centrifuging at 900 x g.
- Place the cleaned eggs on fresh NGM-plates seeded with OP50 bacteria. Grow a bleached population on OP50 bacteria at 15 °C until they reach L4 stage (approximately 4 days). L4 larvae can be recognized by a white patch approximately halfway along the ventral side of the worm using a standard stereomicroscope.
NOTE: To achieve the germ cell to neuron conversion phenotype upon depletion of lin-53, it needs to be depleted in the parental generation (P0). The scoring for the conversion phenotype is undertaken in the following generation (filial generation F1). To achieve the deplete lin-53 already in the P0, L4 animals are subjected to RNAi.
- Manually transfer 50 L4 stage worms per replicate using a platinum wire to an NGM plate that does not contain any bacteria and let worms move away from any transferred OP50 bacteria. Let worms move on the plate for around 5 minutes. Use bacteria from the NGM-Agar RNAi plates previously seeded with lin-53 RNAi bacteria (or empty vector control) to transfer to the respective RNAi plates.
- Avoid transferring OP50 bacteria to the actual RNAi plates. Work fast to minimize exposure time of the strain to temperatures higher than 15 °C.
- Incubate worms on NGM-Agar RNAi plates at 15 °C for approximately 7 days until F1 progeny of the worms reaches L3 – L4 stage. They can be clearly separated from the P0 animals since they are bigger and thicker than the F1 progeny.
- Visually check under a standard stereomicroscope whether F1 progeny worms show the protruding vulva (pvul) phenotype as shown in Figure 1. RNAi against lin-53 has pleiotropic effects and causes the pvul phenotype which can be used to assess whether RNAi against lin-53 has been successful.
NOTE: RNAi against lin-53 can also cause lethality of F1 embryos. This effect increases if plates are exposed to higher degrees than 15 °C before animals reached the L3 – L4 stage. Dead embryos can be recognized by arrested development and lack of hatching. - Incubate plates in the dark since IPTG is light sensitive.
- Visually check under a standard stereomicroscope whether F1 progeny worms show the protruding vulva (pvul) phenotype as shown in Figure 1. RNAi against lin-53 has pleiotropic effects and causes the pvul phenotype which can be used to assess whether RNAi against lin-53 has been successful.
Risultati
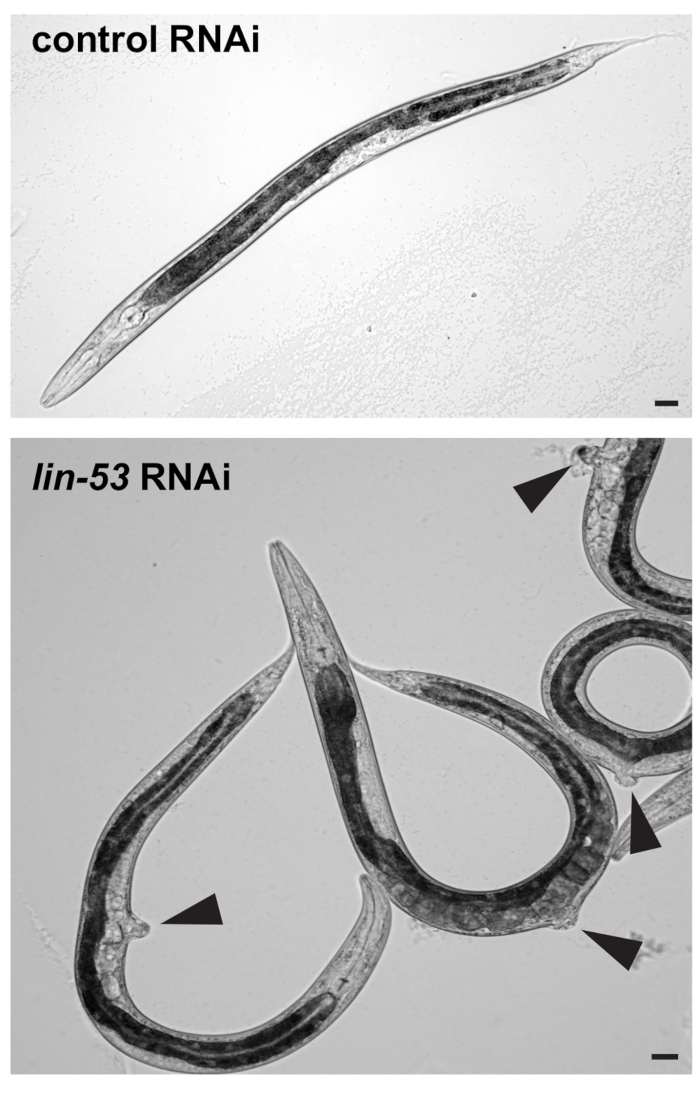
Figure 1: Pvul phenotype caused by RNAi against lin-53. (Top) DIC picture of L4/young adult stage F1 progeny worms derived from control or lin-53 RNAi treated mothers. Scale bars = 20 µm. (Bottom) Animals treated with lin-53 RNAi display the protruding vulva (pvul) phenotype (black arrow-heads). The pvul phenotype confir...
Materiali
| Name | Company | Catalog Number | Comments |
| Chemicals | |||
| Agar-Agar, Kobe I | Roth | 5210.1 | |
| Bactopeptone | A. Hartenstein GmbH | 211 677 | Peptone Media |
| Yeast extract | AppliChem | A1552,0100 | |
| Amphotericin B | USBiological | A2220 | |
| CaCl2 | Merck Biosciences | 208290 | |
| Cholesterol | Roth | 8866.2 | |
| Gelatine | Roth | 4275.3 | |
| IPTG | Sigma | 15502-10G | |
| K2PO4 | Roth | T875.2 | |
| KH2PO4 | Roth | 3904.2 | |
| MgSO4 | VWR | 25,163,364 | |
| Na2HPO4 | Roth | P030.1 | |
| NaCl | Roth | 9265.1 | |
| NaClO | Roth | 9062.3 | |
| NaOH (5 N) | Roth | KK71.1 | |
| Tris | Roth | AE15.2 | |
| Carbenicillin | Roth | 634412 | |
| Tetracycline | Roth | 2371.2 | |
| Incubators | |||
| Incubator | Sanyo MIR-5 | 5534210 | for maintenance of worm strains at 15 °C or 25 °C |
| Microscopes | |||
| Stereomicroscope | SMZ745 | Nikon | |
| Bacterial strains | |||
| Escherichia coli HT115 | F-, mcrA, mcrB, IN(rrnD-rrnE)1, rnc14::Tn10(DE3 lysogen: lavUV5promoter -T7 polymerase) (IPTG-inducible T7 polymerase) (RNAseIII minus). | ||
| Escherichia coli OP50 | Uracil auxotroph E. coli strain | ||
| Worm strains | |||
| BAT28: otIs305 (hsp16.2prom::che-1::3xHA) ntIs1 (gcy-5prom::gfp) V. | derived from OH9846 by 4x times more back crossing with N2 | ||
| RNAi Clones | |||
| lin-53 | SourceBioscience | ID I-4D14 | Ahringer library 16B07 ChromI, K07A1.12 |
| empty vetor: L4440 | Addgene | #1654 |
This article has been published
Video Coming Soon
Source: Kolundzic, E., et al. Application of RNAi and Heat-shock-induced Transcription Factor Expression to Reprogram Germ Cells to Neurons in C. elegans. J. Vis. Exp. (2018).