A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Isolation of High Purity Tissues from Developing Barley Seeds
In This Article
Summary
Here we present a protocol for high purity manual isolation and quality control of embryo, endosperm and seed maternal tissues during entire barley seed development.
Abstract
Understanding the mechanisms regulating the development of cereal seeds is essential for plant breeding and increasing yield. However, the analysis of cereal seeds is challenging owing to the minute size, the liquid character of some tissues, and the tight inter-tissue connections. Here, we demonstrate a detailed protocol for dissection of the embryo, endosperm, and seed maternal tissues at early, middle, and late stages of barley seed development. The protocol is based on a manual tissue dissection using fine-pointed tools and a binocular microscope, followed by ploidy analysis-based purity control. Seed maternal tissues and embryos are diploid, while the endosperm is triploid tissue. This allows the monitoring of sample purity using flow cytometry. Additional measurements revealed the high quality of RNA isolated from such samples and their usability for high-sensitivity analysis. In conclusion, this protocol describes how to practically dissect pure tissues from developing grains of cultivated barley and potentially also other cereals.
Introduction
Seeds are complex structures composed of several tissues of maternal and filial origin1. Cereal grains represent a special type of seed, with the largest part being formed by endosperm, a specialized triploid tissue that protects and nourishes the embryo. Cereals provide around 60% of global food resources and are the most valuable output from plant production2. The knowledge of molecular processes controlling cereal seed development is important due to their economic prominence and central role in plant reproduction1,3.
Cultivated barley (Hordeum vulgare subsp. vulgare; 2n = 2x = 14; 1C = 5.1 Gbp) is the fourth most important cereal crop worldwide. It is used for animal feed, food, and biotechnology4. Besides that, it is also a classical temperate zone cereal crop model species of growing importance5. Barley genomic resources include genetic maps, collections of cultivars, landraces and mutants, high-quality genome assemblies and annotations as well as transcriptomic data of the major developmental stages5,6,7. Also, barley genes are used for genetic improvements of other cereals. Resistance to abiotic stresses such as drought and salinity, specific pathogens, and high content of beneficial compounds (e.g., β-glucan) make barley a valuable source of traits for wheat breeding8.
Seed development is initiated by fertilization on the day of pollination (DOP). DOP is defined by evaluation of the morphology of stigma and anthers according to the Waddington scale (W10.0)9. The spikes containing non-pollinated flowers were characterized by compact (unbranched) stigma and green anthers, whereas pollinated spikes contained extended spiklets, extended and widely branched stigma, swollen ovule, opened anthers and free pollen. The flowers at DOP represented an intermediate phenotype. The anthers had a yellow color, disrupted easily and then released pollen. Stigma had widely spread sigmatic branches of the pistil (Figure 1C).
Barley seed development includes three partially overlapping stages1,10. The stage I (0 – 6 days after pollination; DAP) is launched by double fertilization, typified by cell proliferation and the absence of starch synthesis; stage II (7 – 20 DAP) comprises differentiation and great biomass gain accompanied by the production of starch and protein storage molecules; stage III (after 21 DAP) corresponds to seed maturation, weight reduction by desiccation and the onset of dormancy. Alternatively, the phases are called early, middle and late, respectively11.
Barley grain is covered by hulls, which consist of the lemma, palea, and glumes12. In most barley genotypes, the hulls tightly wrap dry seeds. The seed itself is formed by the embryo, endosperm and seed maternal tissues (Figure 1A). The diploid embryo originates from the fertilization of the egg cell by one sperm cell nucleus. In the fully developed seed, the embryo consists of the embryonic axis with the coleorhiza surrounding the radicle, the coleoptile enclosing the shoot meristem and primary leaves, and the scutellum (cotyledon)1,10,13,14. The triploid endosperm is the result of fertilization of the diploid central cell by the second sperm cell nucleus. The proliferation of endosperm begins with the syncytial (coenocyte) stage, where the dividing nuclei are pushed to the periphery by the central vacuole. At the end of the syncytial phase, microtubules form a radial network around the nuclei and indicate the anticlinal cell wall formation and the onset of endosperm cellularization. Endosperm differentiation occurs simultaneously with the cellularization and results in five major tissues: the starchy endosperm, the transfer cells, the aleurone and subaleurone layers, and the embryo surrounding region. Seed maternal tissues are a multi-layered diploid structure of maternal origin containing pericarp and seed coats10,12. Seed maternal tissues include a nucellar projection on the dorsal side of the grain that has a transport-related function, and becomes embedded in endosperm at later stages of seed development15.
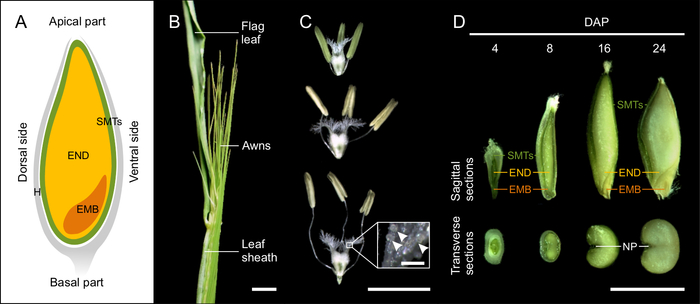
Figure 1: Developing barley seeds. (A) The schematic drawing of cereal grain at the sagittal plan with indicated seed maternal tissues (SMTs, green), endosperm (END, yellow), embryo (EMB, orange) and hulls (H, grey). (B) Morphology of barley spike close to the anthesis. Scale bar = 1 cm. (C) Morphology of stigma and anthers at the stages before, during and after pollination. Inset shows detail of the stigma with pollen grains (arrowheads). Scale bar = 5 mm, inset bar = 200 µm. (D) Sagittal and transverse sections of 4, 8, 16 and 24 DAP seeds. (NP, nucellar projection) Scale bar = 5 mm. Please click here to view a larger version of this figure.
Recent progress in high-throughput genomics provides the tools for the study of individual seed tissue development. However, the major obstacle of this purpose is the compact structure and tight adhesion of the seed tissues1. We developed a protocol for high purity dissection of seed tissues from developing barley seeds with possibility to subsequent use for highly sensitive analyses, such as RNA-sequencing. In addition, the presented protocol can be easily adapted to other cereals.
Protocol
1. Growing plants
NOTE: Considering that a single barley plant usually has 5 to 6 tillers and only the middle 5 to 6 spikelets of each spike should be used for dissection, then a maximum yield per plant is 72 seeds for two-row and 216 seeds for six-row cultivars.
- To germinate barley seeds, prepare a Petri dish padded with three layers of cellulose tissue paper covered with one layer of filter paper. Moisturize it with distilled water, so there is no excess water, put the seeds on the surface and close the Petri dish. Filtration paper avoids growing the roots through the cellulose tissue. Germinate the seeds for 3 days at 25 °C in the dark.
NOTE: Alternatively, germinate seeds by putting them directly in a wet soil mixture (see step 1.2). - Transfer germinated seeds with a visible radicle and shoot of about 5 cm into 5 cm x 5 cm peat pots with a mixture of soil and sand (3:1, v/v). Water regularly. After 10 days, transfer the plants into 12 cm x 12 cm pots filled with the same soil mixture.
- Grow the plants in a climatic chamber under the controlled long-day regime (16 h day 20 °C, 8 h night 16 °C; light intensity 200 µmol m-2 s-1; humidity 60%).
NOTE: Spring barley requires approximately 8-10 weeks from sowing to the beginning of anthesis, with no requirement for vernalization. Winter barley needs 7-8 weeks of vernalization (short-day conditions, 8 h day 4 °C, 16 h night 4 °C; light intensity 200 µmol m-2 s-1; humidity 85%) to induce flowering.
2. Determination of pollination
NOTE: Precise determination of pollination is needed for proper estimation of developmental progression. Barley is a self-pollinating species. To define day of pollination (DOP), we monitored the day of self-pollination. This trait is cultivar specific, but starting protrusion of the awns from the leaf sheath is a good indicator of approaching DOP (Figure 1B).
- Open the leaf-sheath covering the spike. Use fine-pointed tweezers to check anthers and the ovary inside the spikelets in the central part of the spike. Spikelets with yellow anthers and “fluffy” stigma will pollinate within few hours16 and are considered as DOP (Figure 1B).
- Clip off the spike near the tip of the last spikelet, and remove the flag leaf and the upper part of the awns. Then, clip off the top 1/3 of hulls in each spikelet. This dries the anthers and leads to their more synchronized opening and release of pollen.
- Cover the spike with a glassine bag with the spike ID, plant number and defined DOP date. This also prevents cross-pollination, which may compromise specific experiments.
- Note the information to a tabular editor. Use the following formula to calculate the day after pollination (DAP) when tissue isolation should take place.
xDAP = DOP + x
x = expected DAP
NOTE: The values should have ‘Data’ format. - For seed tissues dissection, collect the spikes at DAP according to the prepared tabular calendar.
3. Dissection of the seed tissues
NOTE: The following steps should be performed using a stereomicroscope. Remove the hulls before dissection using tweezers. Note that hulls become drier and more adherent from around 16 DAP. To keep physiological conditions and avoid drying of the plant materials during dissection, moisten the samples by putting them into a drop of 1x PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH = 7.4). Use a new seed for dissection of each tissue to avoid DNA, RNA, or protein degradation due to extended sample collection time. For RNA isolation from dissected material, use only RNase-free materials and chemicals. Do not exceed the total dissection time 15 minutes for one sample consisting typically from tissues dissected from 5-10 seeds to minimize RNA degradation.
- Remove the rest of the hulls using fine-pointed tweezers before tissue dissection. Moisturizing with 1× PBS for 1 minute helps to remove dry residues of the spikelet.
- Place the peeled seed on a Petri dish with a drop of 1× PBS and dissect individual parts using fine-pointed tweezers, fine-needle, and microcapillary pipette. A slightly different strategy is applied for dissection of individual tissues: the seed maternal tissues (step 3.3), the embryo (step 3.4) and the endosperm (step 3.5).
- Dissection of seed maternal tissues
- Dissection from seeds up to 8 DAP
- Place a seed on the dorsal side and gently cut the seed along a longitudinal axis, and peel off seed maternal tissues except the last layer bordering endosperm from the apical to the basal part using tweezers.
- Collect seed maternal tissues from 5 to 10 seeds into a 1.5 mL tube with 50 µL of 1× PBS, discard the buffer using a pipette, rinse the tissue twice with 100 µL of PBS, remove excessive buffer by pipetting and close the tube and freeze in liquid nitrogen or use directly for flow cytometric ploidy measurement. The amount of material is sufficient for typically one downstream application (e.g., RNA isolation or flow cytometric ploidy measurement).
- Dissection from seeds after 8 DAP
- Place a seed on the dorsal side, gently cut in the middle of the ventral side of seed maternal tissues and gradually peel off the tissue around whole seed including nucellar projection. For each downstream application collect and wash the tissue from 5 to 10 seeds as described in step 3.3.1.
- Dissection from seeds up to 8 DAP
- Dissection of embryo
- Dissection from seeds at 8 DAP and younger
- Place a seed on the dorsal side and cut basal 1/3 of the seed. Carefully split separated part in half and release the embryo. For each downstream application collect and wash the embryos from 10 to 20 seeds as described in 3.3.1.
- Dissection from seeds after 8 DAP
- Place a seed on the dorsal side and remove seed maternal tissues from the basal part of the ventral side. Gently disturb the thin layer of endosperm around the perimeter of the embryo by fine-needle or fine-pointed tweezers and peel out the embryo. For each downstream application collect and wash embryos from up to 5 seeds as described in 3.3.1.
- Dissection from seeds at 8 DAP and younger
- Dissection of endosperm
- Dissection of syncytial endosperm from 4 DAP seeds.
- Place a seed on the dorsal side, and remove seed maternal tissues except the last layer of cells bordering endosperm. Gently puncture layer in the middle of the ventral side by a thin needle, and suck the syncytial endosperm by capillary action using a microcapillary pipette.
- For each downstream application collect liquid endosperms from 10 to 15 seeds into a new 1.5 mL tube with buffer suitable for the planned downstream analysis (i.e., 1x PBS, RNA isolation buffer, flow cytometry buffer). Buffer volume should reflect the protocol for the planned downstream aplication. Freeze in liquid nitrogen.
- Dissection of celullarizing endosperm from 5 to 8 DAP seeds
- Place a seed on the dorsal side, and remove all seed maternal tissues and embryo. For each downstream application collect and wash the endosperm from 10 to 15 seeds into a new 1.5 mL tube with 1x PBS and freeze in liquid nitrogen.
- Dissection of celullarized endosperm from seed after 8 DAP
- Place a seed on the dorsal side, remove all seed maternal tissues and embryo. For each downstream application collect and wash endosperm from a single seed per tube as described in step 3.3.1.
- Dissection of syncytial endosperm from 4 DAP seeds.
- Store the tubes with isolated material at – 80 °C until use.
NOTE: The protocol can be paused here.
4. Control of tissue purity using flow cytometry
NOTE: The sample purity can be checked using flow cytometry before RNA isolation. Proper instrument calibration is critical for the biological sample analysis. The flow cytometer/ploidy analyzer optics should be adjusted using calibration beads (fluorescently stained polystyrene microspheres highly uniform with respect to their size and fluorescence intensity) until the maximal peak sharpness, typically reaching the coefficient of variation (CV) < 2%. Cereal seed tissues contain mainly populations of G1, G2 and endoreduplicated nuclei; therefore, using a logarithmic scale is recommended. Start with a leaf tissue that contains mostly G1 nuclei and serves as a basal ploidy control.
- Use freshly prepared samples kept on ice (see step 3.3.1) or frozen tissue as described17.
NOTE: Because the whole < 8 DAP sample is used for flow cytometry, this represents only an indirect control. We recommend researchers performing multiple isolations and measurements until reaching high proportion of pure samples (>90%) before proceeding to RNA isolation with < 8 DAP samples. - Release the nuclei from the 4 and 8 DAP embryo samples (for other samples see step 4.4) by homogenizing the tissues by 5 to 10 turns of the plastic pestle in 1.5 mL tube containing 300 µL of Otto I buffer (0.1 M citric acid monohydrate, 0.5% (v/v) Tween 20, filtered through a 0.22 µm filter)18.
- Filter the crude suspension through 50 µm nylon mesh into a flow-cytometry analysis tube and add 600 µL of Otto II buffer (0.4 M Na2HPO4·12H2O) containing 2 µg mL-1 DAPI (4´,6-diamidino-2-phenylindole)18 to stain DNA.
- Place all other tissues (including 16 DAP or older embryos) on a Petri dish containing 500 µL of Otto I buffer and homogenized by chopping with a razor blade. Filter the suspension as in step 4.3 and stain with 1 mL of Otto II buffer containing DAPI.
NOTE: Manipulation with a sharp double edge razor blade requires special attention. To reduce the risk of injury, there are a single edge razor blades or special blade holders available. - Estimate the nuclear DNA content of the sample using a flow cytometer. At least 2000 particles per sample are required for analyzing the sample purity.
5. RNA isolation and quality measurement
- Use frozen tissue to prevent RNA degradation by endogenous ribonucleases. From seed maternal tissues and embryo samples isolate RNA using commercially available kits or TRIzol reagent19. Due to a high starch content in endosperm tissues, isolate total RNA from all samples using commercial on-column RNA extraction protocols for problematic tissues (e.g., Spectrum Plant Total RNA Kit) with an on-column DNase I treatment20.
- Measure RNA concentration and integrity using a dedicated protocol for RNA gel electrophoresis or Agilent 2100 Bioanalyzer.
NOTE: Intact total RNA has a clear 18S and 25S rRNA bands/peaks of size around 1.9 and 3.7 kb respectively. The 25S rRNA band should be approximately two times more intense than the 18S rRNA band.
Results
To perform a tissue-specific transcriptomic analysis of barley seed development, we established a protocol for high purity tissue isolation. The protocol is based on the manual dissection of embryo, endosperm and seed maternal tissues from peeled (after manual hull removal) grains (Figure 1A). The protocol was successfully used for isolating materials from several two- and six-row spring barley cultivars, and the spikes were harvested at a given DAP and directly used for extraction without f...
Discussion
Here, we present a protocol that allows high purity isolation of barley seed tissues. Although it was developed and tested for barley, it can be easily adopted to other members of the Triticeae tribe such as wheat, oat, rye or triticale27. The initial part of the protocol, focusing on seed tissue dissection, does not require any non-standard or expensive equipment and therefore should be accessible to many scientists. A highly specialized instrument such as a flow cytometer is required fo...
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Jan Vrána and Dr. Mahmoud Said for the maintenance of flow cytometers, Eva Jahnová for preparation of buffers Marie Seifertová for list of materials and Zdenka Bursová for plant care. This work was supported primarily from the Czech Science Foundation grant 18-12197S. A.P. was further supported by the J. E. Purkyně Fellowship from the Czech Academy of Sciences and the ERDF project "Plants as a tool for sustainable global development" (No. CZ.02.1.01/0.0/0.0/16_019/0000827).
Materials
| Name | Company | Catalog Number | Comments |
| 0.22 um filter | Merck | SLGSV255F | |
| 1.5 ml Eppendorf tube | Sarstedt | 72.690.001 | |
| 4',6-diamididno-2-phenylindole | Invitrogen | D21490 | |
| 50 um nylon mesh | Silk a Progrers | uhelon 120 T | |
| Agilent 2100 Bioanalyzer | Agilent | G2939BA | |
| Bulb Assembly | Drummond Scientific Company | 1-000-9000 | |
| Calibration beads | Invitrogen | A16502 | |
| Cellulose tissue paper | |||
| Citric acid monohydrate | Penta | 13830-31000 | |
| Climatic chamber | Weiss Gallenkamp | ||
| DNase I | Sigma Aldrich | DNASE70 | |
| Filter paper | Fagron | ||
| Fine-pointed tweezers | Fine Science Tools | 11254-20 | |
| Flow cytometer | Sysmex-Partec | ||
| Flow cytometry tube | Sarstedt | 55.484 | |
| Freezer | |||
| Glassine bag | |||
| KCl | Lachner | 30076-AP0 | |
| KH2PO4 | Litolab | 100109 | |
| Liquid nitrogen | Linde | ||
| Microcapillary pipette | Fivephoton Biochemicals | MGM 1C-20-30 | |
| Minutien Pins | Fine Science Tools | 26002-20 | |
| Na2HPO4 | Lachema | ||
| Na2HPO4.12H2O | Lachner | 30061-AP0 | |
| NaCl | Lachner | 30093-AP0 | |
| Peat pots | Jiffy | 5x5 cm | |
| Petri dish | |||
| Pin Holder | Fine Science Tools | 26016-12 | |
| Plastic pestle | p-Lab | A199001 | |
| Pots | 12x12 cm | ||
| Razor blade | Gillette | ||
| RNAse zap | Invitrogen | AM9780 | |
| Sand | |||
| Scissors | Fine Science Tools | 14060-11 | |
| Soil | |||
| Spectrum Plant Total RNA Kit | Sigma Aldrich | STRN50 | |
| Stereomicroscope | Olympus | ||
| Tween 20 | Sigma Aldrich | P2287 | |
| TRIzol reagent | Invitrogen | 15596026 | |
| RNA 6000 Pico Kit | Agilent | 5067-1513 |
References
- Sreenivasulu, N., et al. Barley grain development: Toward an integrative view. International Review of Cell and Molecular Biology. 281, 49-89 (2010).
- Baik, B. K., Ullrich, S. E. Barley for food: Characteristics, improvement, and renewed interest. Journal of Cereal Science. 48 (2), 233-242 (2008).
- Langridge, P., Stein, N., Muehlbauer, G. J. Economic and Academic Importance of Barley. The Barley Genome. , 1-10 (2018).
- Mascher, M., et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 544 (7651), 427-433 (2017).
- Pseudomolecules and annotation of the second version of the reference genome sequence assembly of barley cv. Morex V2. Morex Available from: https://edal.ipk-gatersleben.de (2019)
- Rapazote-Flores, P., et al. BaRTv1.0: An improved barley reference transcript dataset to determine accurate changes in the barley transcriptome using RNA-seq. BMC Genomics. 20 (1), 968 (2019).
- Molnár-Láng, M., Linc, G., Szakács, &. #. 2. 0. 1. ;. Wheat-barley hybridization: The last 40 years. Euphytica. 195 (3), 315-329 (2014).
- Waddington, S. R., Cartwright, P. M., Wall, P. C. A quantitative scale of spike initial and pistil development in barley and wheat. Annals of Botany. 51 (1), 119-130 (1983).
- Sabelli, P. A., Larkins, B. A. The development of endosperm in grasses. Plant Physiology. 149 (1), 14-26 (2009).
- Dante, R. A., Larkins, B. A., Sabelli, P. A. Cell cycle control and seed development. Frontiers in Plant Science. 5, 1-14 (2014).
- Rodríguez, M. V., Barrero, J. M., Corbineau, F., Gubler, F., Benech-Arnold, R. L. Dormancy in cereals (not too much, not so little): About the mechanisms behind this trait. Seed Science Research. 25 (2), 99-119 (2015).
- Sreenivasulu, N., et al. Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds. Plant Journal. 47 (2), 310-327 (2006).
- Olsen, O. A. Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell. 16, 214-227 (2004).
- Thiel, J., et al. Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells: A microdissection-based transcriptome study of young barley grains. Plant Physiology. 148 (3), 1436-1452 (2008).
- Weschke, W., et al. Sucrose transport into barley seeds: Molecular characterization of two transporters and implications for seed development and starch accumulation. Plant Journal. 21 (5), 455-467 (2000).
- Staszak, A. M., Rewers, M., Sliwinska, E., Klupczyńska, E. A., Pawłowski, T. A. DNA synthesis pattern, proteome, and ABA and GA signalling in developing seeds of Norway maple (Acer platanoides). Functional Plant Biology. 46 (2), 152-164 (2019).
- Otto, F. Chapter 11 DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods in Cell Biology. 33, 105-110 (1990).
- Fisher Scientific. . Procedural guidelines. , (2020).
- Spectrum TM. . Plant Total RNA Kit. , (2020).
- Nowicka, A., et al. Dynamics of endoreduplication in developing barley seeds. Journal of Experimental Botany. , eraa453 (2020).
- Chen, J., et al. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiology. 166 (1), 252-264 (2014).
- Zhang, S., Laurie, A. E., Ae, W., Meng, L., Lemaux, P. G. Similarity of expression patterns of knotted1 and ZmLEC1 during somatic and zygotic embryogenesis in maize (Zea mays L.). Springer. 215 (2), 191-194 (2002).
- Doll, N. M., et al. Transcriptomics at maize embryo/endosperm interfaces identifies a transcriptionally distinct endosperm subdomain adjacent to the embryo scutellum. Planta. 32 (4), 833-852 (2020).
- Yi, F., et al. High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell. 31 (5), 974-992 (2019).
- Gómez, E., et al. The maize transcription factor myb-related protein-1 is a key regulator of the differentiation of transfer cells. Plant Cell. 21 (7), 2022-2035 (2009).
- Bewley, J. D., Black, M., Halmer, P. The encyclopaedia of seeds: Science, technology and uses. CABI. , (2006).
- Liew, L. C., et al. Temporal tissue-specific regulation of transcriptomes during barley (Hordeum vulgare) seed germination. Plant Journal. 101 (3), 700-715 (2020).
- Pirrello, J., et al. Transcriptome profiling of sorted endoreduplicated nuclei from tomato fruits: how the global shift in expression ascribed to DNA ploidy influences RNA-Seq data normalization and interpretation. Plant Journal. 93 (2), 387-398 (2018).
- Sreenivasulu, N., et al. Transcript profiles and deduced changes of metabolic pathways in maternal and filial tissues of developing barley grains. Plant Journal. 37 (4), 539-553 (2004).
- Bian, J., et al. Transcriptional dynamics of grain development in barley (Hordeum vulgare L). International Journal of Molecular Sciences. 20 (4), 962 (2019).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved