Для просмотра этого контента требуется подписка на Jove Войдите в систему или начните бесплатную пробную версию.
Method Article
Экспертиза BCL-2 Семья функции с большим однослойные везикулы
В этой статье
Резюме
Biochemically-defined large unilamellar vesicles (LUVs) are a convenient model system to analyze BCL-2 family interactions with immediate implications in better understanding the mitochondrial pathway of apoptosis. A method to produce LUVs, along with standard BCL-2 family protein combinations and controls to examine LUV permeabilization, are presented.
Аннотация
The BCL-2 (B cell CLL/Lymphoma) family is comprised of approximately twenty proteins that collaborate to either maintain cell survival or initiate apoptosis1. Following cellular stress (e.g., DNA damage), the pro-apoptotic BCL-2 family effectors BAK (BCL-2 antagonistic killer 1) and/or BAX (BCL-2 associated X protein) become activated and compromise the integrity of the outer mitochondrial membrane (OMM), though the process referred to as mitochondrial outer membrane permeabilization (MOMP)1. After MOMP occurs, pro-apoptotic proteins (e.g., cytochrome c) gain access to the cytoplasm, promote caspase activation, and apoptosis rapidly ensues2.
In order for BAK/BAX to induce MOMP, they require transient interactions with members of another pro-apoptotic subset of the BCL-2 family, the BCL-2 homology domain 3 (BH3)-only proteins, such as BID (BH3-interacting domain agonist)3-6. Anti-apoptotic BCL-2 family proteins (e.g., BCL-2 related gene, long isoform, BCL-xL; myeloid cell leukemia 1, MCL-1) regulate cellular survival by tightly controlling the interactions between BAK/BAX and the BH3-only proteins capable of directly inducing BAK/BAX activation7,8. In addition, anti-apoptotic BCL-2 protein availability is also dictated by sensitizer/de-repressor BH3-only proteins, such as BAD (BCL-2 antagonist of cell death) or PUMA (p53 upregulated modulator of apoptosis), which bind and inhibit anti-apoptotic members7,9. As most of the anti-apoptotic BCL-2 repertoire is localized to the OMM, the cellular decision to maintain survival or induce MOMP is dictated by multiple BCL-2 family interactions at this membrane.
Large unilamellar vesicles (LUVs) are a biochemical model to explore relationships between BCL-2 family interactions and membrane permeabilization10. LUVs are comprised of defined lipids that are assembled in ratios identified in lipid composition studies from solvent extracted Xenopus mitochondria (46.5% phosphatidylcholine, 28.5% phosphatidylethanoloamine, 9% phosphatidylinositol, 9% phosphatidylserine, and 7% cardiolipin)10. This is a convenient model system to directly explore BCL-2 family function because the protein and lipid components are completely defined and tractable, which is not always the case with primary mitochondria. While cardiolipin is not usually this high throughout the OMM, this model does faithfully mimic the OMM to promote BCL-2 family function. Furthermore, a more recent modification of the above protocol allows for kinetic analyses of protein interactions and real-time measurements of membrane permeabilization, which is based on LUVs containing a polyanionic dye (ANTS: 8-aminonaphthalene-1,3,6-trisulfonic acid) and cationic quencher (DPX: p-xylene-bis-pyridinium bromide)11. As the LUVs permeabilize, ANTS and DPX diffuse apart, and a gain in fluorescence is detected. Here, commonly used recombinant BCL-2 family protein combinations and controls using the LUVs containing ANTS/DPX are described.
протокол
1. Combine Lipids and Create a Lipid Film
- Working in a fume hood with minimal lighting, chloroform-solubilized lipids are combined into a 1.5 ml amber glass vial using a chloroform-rinsed Hamilton gas-tight glass syringe. This combination will yield approximately 4 mg of lipid (Figure 1A).
- Cardiolipin 10 mg/ml (CL) - 63 μl
- Phosphatidylcholine 10 mg/ml (PC) - 84 μl
- Phosphatidylethanoloamine 10 mg/ml (PE) - 125 μl
- Phosphatidylinositol 10 mg/ml (PI) - 59.5 μl
- Phosphatidylserine 10 mg/ml (PS) - 65 μl
- To remove the chloroform, apply a gentle stream of an inert gas (e.g., nitrogen or argon) into the vial. A lipid film will become visible on the bottom and sides of the vial when dry, which typically takes 2 - 3 hours (Figure 1A).
2. Prepare Liposome Solution
- In 1.7 ml microcentrifuge tubes, prepare 500 μl of 12.5 mM ANTS in LUV buffer (0.2 mM EDTA, 10 mM HEPES [pH 7], 200 mM KCl, 5 mM MgCl2 ) and 500 μl of 45 mM DPX in LUV buffer (Figure 1B).
- Combine the ANTS and DPX solutions, vortex for 1 min, and sonicate for 1 min at room temperature. Sonication is done in an ultrasonic water bath (e.g., Branson 3510MTH, no setting, just 'on'). This ensures the solutions are entirely solubilized and homogeneous.
- Add the ANTS/DPX solution to the lipid film, and seal the vial with parafilm (Figure 1B).
- Sonicate the lipid film in the above water bath for 5 min. The solution will appear milky after sonication. Ensure the lipid film is completely solubilized as small lipid aggregates will negatively impact on proceeding steps. Also, check the vial's sides and bottom to ensure there is no remaining lipid film (Figure 1B).
3. Extrusion and Purification of Liposomes
- Assemble the extruder according to the manufacturer's guidelines using LUV buffer to moisten the filter supports and polycarbonate membrane. Tighten the unit's casing so that there will be no loss of volume during the extrusion, but not overly tight as to rupture the membrane. It's also wise to test the extruder with LUV buffer to ensure there are no leaks or problems.
- Start with the lipid solution in the right syringe, extrude the lipid solution 31 times, and the LUVs will finish in the left syringe; this ensures all the lipids have passed through the membrane at least once. The first few times may required a little more pressure to pass through the membrane. If the solution suddenly passes through the membrane quickly, check the membrane for rupture and replace, if necessary (Figure 1C).
- Set-up a 10 ml Sepharose S-500 gravity flow column (1.0 cm x 20 cm, 10 ml bead volume). Wash with 3 column volumes of LUV buffer, allow the column flow to stop, and cap. Since this step takes a while, it is preferred to start the washing when the lipids are combined to dry. Just keep the column capped until you are ready to use it.
- Add the extruded LUVs to the column in one step, avoid disrupting the bead interface. Allow the buffer to run out of the column, the flow will stop, and plug. Gently fill the column with LUV buffer. Again, use caution when adding buffer so that the beads are not disturbed (Figure 1C).
- Remove the plug and collect 6 x 1 ml fractions. The LUVs normally elute in fractions 3 and 4, and can be identified by a cloudy appearance. Once the fractions are collected, wash the column with 5 volumes of water, and 2 volumes of 20% ethanol. Store the column at 4 °C (Figure 1C).
- Store the fractions at 4 °C in the dark until tested.
4. Test Column Fractions
- The column fractions are now tested for fluorescence comparing LUV buffer and LUV buffer supplemented with 0.5% CHAPS. Fractions that increase in fluorescence due to the presence CHAPS contain LUVs. Pipette 100 μl of LUV buffer or LUV buffer + 0.5% CHAPS into six wells each, and add 5 μl of the appropriate fraction, mix. Use opaque, black, untreated, 96 well plates for all LUV studies (Figure 1D).
- Read the plate at 37 °C with the following parameters: Excitation wavelength: 355 nm; Emission wavelength: 520 nm; Gain (voltage): 125; Optics position: Top; Read height: 5.5 mm. This set-up is for the BioTek H1 Synergy using Gen 5 2.0 software (Figure 1D).
- Fractions 3 and 4 generally show a 5 - 10 fold increase in fluorescence. Once analyzed, combine the LUV-containing fractions and store at 4 °C in the dark; approximately 2 ml of ~ 2 mM lipid will be purified (Figure 1D).
5. BCL-2 Family Studies Using LUVs
- BCL-2 proteins are tested for their ability to directly induce LUV permeabilization, and for the regulation of permeabilization activity. LUV assays are normally performed in 100 μl volumes, at 37 °C, and can be analyzed as either a single time point after 30 - 60 min (Figures 2A-C, 3A-B), or via kinetic studies every 1 - 2 min for up to 1 hr (Figure 2D). A representative assay design describing components, stock/final concentrations, and order of addition, is provided (Table 1). It is also advisable to have the microplate reader gently shake the plate prior to each reading. The first few min often give sporadic readings (likely due to temperature changes, mixing, etc...), so we generally consider data points after 2 - 3 readings in a kinetic study. It is also not uncommon to generate large error bars for some of the conditions due to variability in LUV batches, and the efficiency of protein interactions at borderline concentrations of activation or inhibition.
- BCL-2 family protein stocks should be diluted into LUV buffer at a concentration of 100x, and necessary titrations of BAX, BID, and all other proteins are performed to find the lowest concentrations necessary to detect minimal BAX-dependent LUV permeabilization in the absence of BID, and significant synergy between BAX and BID. Furthermore, all other proteins require titrations to reduce protein waste, and minimize the concentrations of subsequent protein additions.
6. Representative Results
There are a myriad of options to witness and examine BCL-2 family dependent regulation of LUV permeabilization. To generate a LUV permeabilization positive control, we use detergent-activated BAX12. The detergent, n-octyl-β-D-glucoside (OG), artificially triggers BAX activation (for 100 μl of 2.3 μM OG-BAX: 5 μg BAX + 0.7% OG in LUV buffer, incubate for 60 min at 4 °C, aliquot and store at -80 °C), and therefore OG-BAX (10 - 100 nM) can be used as a reliable positive control. Of note, OG concentrations below 0.025% do not directly affect LUV permeabilization. A 0.5% CHAPS treatment is used to determine the maximum amount of LUV fluorescence per assay, and this value sets the 100% value. All treatments should be set-up in triplicate. An example LUV control assay set-up is shown in Table 1, and corresponding data are presented in Figure 2A.
A necessary starting point for studying LUV permeabilization with BCL-2 family proteins is to establish the concentrations of BAX and BID to promote optimal release. As individual protein preparations will demonstrate different background permeabilization activities, it is advisable to titrate before setting up more sophisticated analyses. BAX (50 - 200 nM) and BID (0 - 10 nM) must be titrated for minimal background release, and optimal synergy, as shown in Figures 2B-C, respectively. Various forms of BID protein can be purchased from R&D Systems; alternatively, the BID BH3 domain peptide is available from Anaspec. There is no commercial form of BAX that is appropriate for LUV assays, therefore it must be expressed and purified using published methods3,13.
Once BAX and BID concentrations and synergy are established, a common assay is to determine the influence of anti-apoptotic proteins on BAX and BID-dependent LUV permeabilization. BCL-xL is an anti-apoptotic BCL-2 member that is commercially available, and in a dose-dependent manner, inhibits the activity of BAX and BID usually in the range of 5 - 10 fold molar excess of BAX or BID (Figure 3A). This inhibitory effect can be reversed by the addition of subsequent BH3-only proteins (2 - 5 fold molar excess to BCL-xL) or BH3 domain peptides (5 - 10 fold molar excess to BCL-xL), which bind to BCL-xL, thus allowing for BAX and BID to synergize and permeabilize the LUVs (Figure 3B). In Figure 3B, we show the effect of full-length recombinant human PUMAβ, and the PUMA BH3 domain peptide is functionally similar (data not shown)8. This experiment is commonly referred to as a 'de-repression assay', as the PUMA protein relieves the inhibitory effect of BCL-xL on BAX and BID. Ideally, the de-repressing BH3-only protein/peptide addition should have minimal effects on BAX activation14.
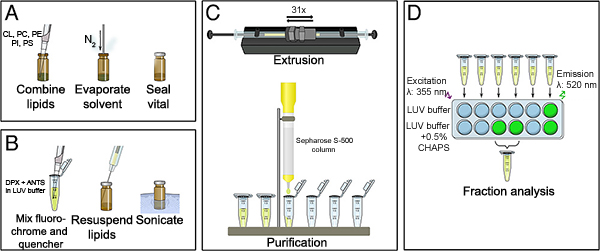
Figure 1. (A) Chloroform solubilized lipids are combined, dried to create a lipid film, and capped. (B) ANTS and DPX solutions are prepared, combined, and sonicated to ensure complete solubilization and homogenization. The solution is added to the lipid film, the vial is sealed with parafilm, and sonicated for 5 min in a water bath. The solution will now appear milky. (C) The sonicated lipid solution is loaded into the mini-extruder, processed 31 times, and added to a prepared Sepharose S-500 column for subsequent purification. (D) Fractions obtained after size exclusion chromatography are analyzed by comparing the relative fluorescent units (RFUs) in LUV buffer and LUV buffer + 0.5% CHAPS. The "LUV buffer" reading indicates background fluorescence with intact LUVs; the "LUV buffer + 0.5% CHAPS" condition will show lysis induced gain of fluorescence, indicative of LUVs. Fractions 3 and 4 usually contain the LUVs, and are combined after analysis. Fractions 5 and 6 contain unincorporated ANTS (i.e., not within the LUVs following extrusion), which show high fluorescence independent of CHAPS. Click here to view larger figure.
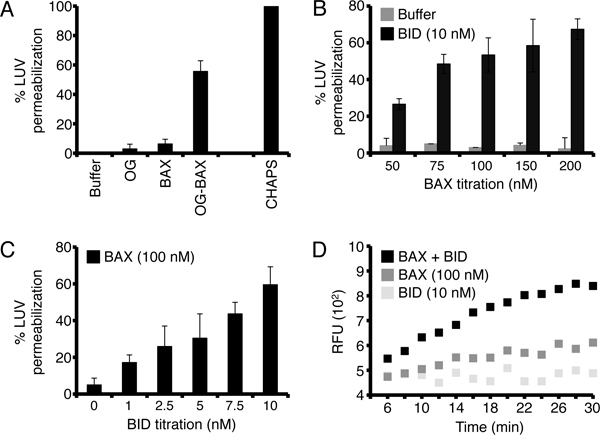
Figure 2. BCL-2 family control experiments and BAX/BID titrations using LUVs. (A) Control experiments. LUVs were treated with LUV buffer alone, 0.025% octylglucoside "OG", 50 nM BAX, or 50 nM OG-BAX. LUVs were incubated for 60 min at 37 °C. The percent (%) permeabilization is determined by subtracting the fluorescence in the buffer treatment from all the samples, 100% permeabilization is determined by a 0.5% CHAPS solubilized sample; divide the experimental values by the CHAPS value to obtain % LUV permeabilization. OG-BAX is detergent-activated BAX, which is a reliable positive control for LUV permeabilization. (B) Determine the optimal BAX concentration. LUVs were treated with increasing concentrations of BAX (50 - 200 nM) in the presence of 10 nM C8-BID "BID", or buffer alone. (C) Determine the optimal BID concentration. LUVs were treated with increasing concentrations of BID (0 - 10 nM) in the presence of 100 nM BAX. (D) An example of a LUV permeabilization kinetic dataset. LUVs were treated with BAX (100 nM) ± BID (10 nM). Readings were performed every two min for 30 min, with two sec of mild plate shaking prior to each read. The first three readings (0, 2, & 4 min) are not included in the graph. The minimum and maximum relative fluorescent units (RFU) were determined as described in section 4.3. The error bars in Figures 2A-C represent the standard deviation from triplicate data. Data in 2D represent a duplicate assay.
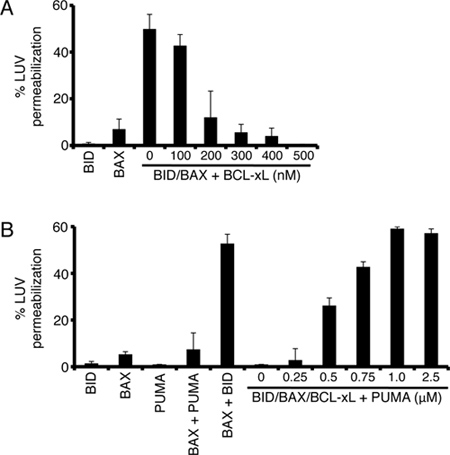
Figure 3. Examples of common BCL-2 family interactions explored using LUVs. (A) BAX and BID mediated LUV permeabilization is inhibited by BCL-xL. LUVs were incubated with 100 nM BAX, 10 nM BID, and increasing concentrations of BCL-xL (0 - 500 nM). LUVs were incubated for 60 min at 37 °C. (B) PUMA de-represses BCL-xL to promote BAX and BID function. LUVs were incubated with combinations of 100 nM BAX, 10 nM BID, 500 nM BCL-xL, and (0 - 2.5 μM) PUMA for 60 min at 37 °C. PUMA was used at 2.5 μM with BAX alone. The error bars represent the standard deviation from triplicate data.
| Order of addition: | 1 | 2 | 3 | 4 | 5 | 6 | |
| CONDITIONS | LUV Buffer | OG (0.7%) | BAX (2.3 μM) | OG-BAX (2.3 μM) | CHAPS (10%) | LUVs (2 mM) | TOTAL |
| Buffer | 95 μl | 5 μl | 100 μl | ||||
| OG (0.015%) | 92.83 μl | 2.17 μl | 5 μl | 100 μl | |||
| BAX (50 nM) | 92.83 μl | - | 2.17 μl | 5 μl | 100 μl | ||
| OG-BAX (50 nM) | 92.83 μl | 2.17 μl | 5 μl | 100 μl | |||
| CHAPS (0.5%) | 90 μl | 5 μl | 5 μl | 100 μl |
Table 1. A control assay example to determine the effect of OG-BAX on LUV permeabilization.
Обсуждение
The described method for generating LUVs enables a rapid and efficient means to test the function of various BCL-2 family proteins, peptides, and related reagents in a biochemically-defined membrane environment similar to the OMM. If using end point values to determine LUV permeabilization, multiple plates can be set-up to analyze hundreds of conditions within a single day. We find that the limiting reagents in these assays tend to be the quality and quantity of recombinant proteins, so dedicating sufficient time and res...
Раскрытие информации
No conflicts of interest declared.
Благодарности
We would like to thank all members of the Chipuk Laboratory for their colleagueship and support. In addition, we would like to give appreciation to Tomomi Kuwana and Donald Newmeyer for developing the experimental foundation for this work. This work was supported by: NIH CA157740 (to J.E.C.), and a pilot project from NIH P20AA017067 (to J.E.C.). This work was also supported in part by a Research Grant 5-FY11-74 from the March of Dimes Foundation (to J.E.C.).
Материалы
| Name | Company | Catalog Number | Comments |
| 1,2-Dioleoyl-sn-Glycero-3-Phosph–thanolamine "PE" | Avanti | 850725C | |
| L-α-Phosphatidylcholine (Egg, Chicken) "PC" | Avanti | 840051C | |
| L-α-Phosphatidylinositol (Liver, Bovine) "PI" | Avanti | 840042C | |
| L-α-Phosphatidylserine (Brain, Porcine) "PS" | Avanti | 8400320 | |
| Cardiolipin (Heart, Bovine - Sodium Salt) "CL" | Avanti | 840012C | |
| Mini-extruder set | Avanti | 610023 | |
| PC membrane 0.2 mM | Avanti | 610006 | |
| Costar black 96 well plate | Fisher Scientific | 07-200-590 | |
| Caspase-8 cleaved human BID | R&D Systems | 882-B8-050 | |
| Human BCL-xL (minus C-terminus) | R&D Systems | 894-BX-050 | |
| BID/PUMA BH3 domain peptides | Anaspec | 61711/62404 | |
| Synergy H1 hybrid multi-mode microplate reader | BioTek | None |
Ссылки
- Chipuk, J. E., Moldoveanu, T., Llambi, F., Parsons, M. J., Green, D. R. The BCL-2 family reunion. Mol. Cell. 37, 299-310 (2010).
- Green, D. R. Apoptotic pathways: ten minutes to dead. Cell. 121, 671-674 (2005).
- Gavathiotis, E. BAX activation is initiated at a novel interaction site. Nature. 455, 1076-1081 (2008).
- Dewson, G. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol. Cell. 30, 369-380 (2008).
- Dewson, G. Bak activation for apoptosis involves oligomerization of dimers via their alpha6 helices. Mol. Cell. 36, 696-703 (2009).
- Gavathiotis, E., Reyna, D. E., Davis, M. L., Bird, G. H., Walensky, L. D. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol. Cell. 40, 481-492 (2010).
- Kuwana, T. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell. 17, 525-535 (2005).
- Chipuk, J. E. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 20327-20332 (2008).
- Letai, A. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2, 183-192 (2002).
- Kuwana, T. lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 111, 331-342 (2002).
- Lovell, J. F. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 135, 1074-1084 (2008).
- Hsu, Y. T., Youle, R. J. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem. 272, 13829-13834 (1997).
- Suzuki, M., Youle, R. J., Tjandra, N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 103, 645-654 (2000).
- Du, H. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem. 286, 491-501 (2011).
Перепечатки и разрешения
Запросить разрешение на использование текста или рисунков этого JoVE статьи
Запросить разрешениеThis article has been published
Video Coming Soon
Авторские права © 2025 MyJoVE Corporation. Все права защищены