A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Analysis of Gene Expression in Emerald Ash Borer (Agrilus planipennis) Using Quantitative Real Time-PCR
In This Article
Summary
Quantitative real-time PCR (qRT-PCR) is an effective tool to diagnose mRNA levels in different insect tissues and developmental stages. In this report we show the use of qRT-PCR to ascertain mRNA levels in different larval tissues and developmental stages of the invasive insect species, emerald ash borer.
Abstract
Protocol
The complete protocol with different steps is depicted in the flow chart in Figure 1. Individual steps constituting dissection, RNA extraction, First Strand cDNA synthesis and qRT-PCR are detailed below.
I. Larval Dissection
- Prior to the start of this procedure, place Emerald ash borer, or EAB, larvae on moist tissue paper until dissections are performed. Keep freshly prepared 1X phosphate buffered saline, or PBS, on ice to keep the buffer cold throughout the dissection.
- To begin, fix the EAB larvae on the dissection plate with fine dissection pins. Then, add cold 1X PBS to the dissection plate.
- First, decapitate larvae with a fine pair of dissection scissors. Subsequently remove the last abdominal segment of the larvae.
- Using a watchmaker pair of forceps, lift the cuticle from the larval carcass. Then, make an incision along the length of the carcass using a pair of scissors. Take care not to rupture the gut as the incision is made.
- Carefully isolate the midgut tissue from other tissues such as fat bodies and connective tissue. Then, rinse the tissue in fresh 1X PBS buffer to ensure that there is no contamination of fatbody. Transfer the isolated midgut to 500 μL of pre-chilled Trizol reagent in a 1.5 mL eppendorf tube.
- Next, isolate the fat body tissue using forceps and transfer it into a 1.5 mL eppendorf tube containing 500 μL of chilled Trizol reagent.
- Finally, isolate the cuticle tissue from the larval carcass by scrapping off any adhering tissue, taking care not to damage tissue integrity. Rinse the isolated cuticle tissue in fresh PBS buffer and transfer it to a 1.5 mL eppendorf tube with 500 μL of chilled Trizol reagent.
- The isolated fat body, cuticle and midgut tissues can be stored at -80 °C until RNA extraction.
- In preparation for RNA extraction, sort the various EAB samples according to the developmental stages. These stages include 1st-, 2nd-, 3rd-, and 4th-instars, prepupae and adults .
II. RNA Extraction (As per manufacturer s protocol for using Trizol Reagent)
Homogenization
- To begin RNA Extraction, first use plastic pestles to homogenize the tissues in the Trizol containing 1.5 mL eppendorf tubes. After homogenization, bring the total volume of each sample to1 mL with the Trizol reagent.
- For the developmental stages, homogenize whole animal in liquid nitrogen with a mortar and pestle. Then add an aliquot of the homogenate (50 μg) to 1 mL of Trizol.
Phase separation:
- Incubate the sample tubes with 1 mL Trizol for 15 minutes at room temperature.
- Following incubation, add 200μL of chloroform to each tube. Immediately after adding chloroform, shake the tubes vigorously for about 15 seconds. Then, incubate the tubes at room temperature for an additional 2-3 minutes.
- Next, centrifuge the samples at 12,000Xg for 15 minutes at 2 °C. After centrifugation, the RNA will be in the aqueous phase. The volume of the aqueous phase should be 600 μl, or 60% of the total Trizol volume.
RNA precipitation
- Carefully remove only the aqueous phase. Presence of substances below the aqueous phase will cause contamination of the extracted RNA. Then, transfer the aqueous phase into an appropriately labeled 1.5 mL eppendorf tube.
- To precipitate the RNA from the aqueous phase, mix 0.5 mL of isopropyl alcohol to each tube. Incubate the samples at room temperature for 10 minutes. After the incubation, centrifuge the tubes at 12,000Xg for 10 minutes at 2°C.
RNA Wash
- Following centrifugation, discard the supernatant from the tubes. RNA is present in the gel-like pellet formed at the bottom of the tube. Wash the pellet with 75% ethanol, adding at least 1 mL of 75% ethanol per 1 mL of Trizol. Mix by vortexing. Then, centrifuge the tubes at 7500Xg for 5 minutes at 2 °C.
RNA elution
- Discard the supernatant from the tubes. Centrifuge the tubes again in the small table centrifuge for 1 minute. Remove any excess supernatant from the tube by careful pipetting. Leave the tube open and let the pellet air dry for 5-10 minutes. Be careful not to over dry the pellet as this will significantly decrease its solubility.
- After air-drying, resuspend the pellet in diethylpyrocarbonate, or DEPC, treated water. The amount of water used to resuspend the pellet will depend on the size of the pellet. Use 50 μL of DEPC treated water for a small pellet or 100 μL for a big pellet. Let the pellet completely dissolve in DEPC treated water by moving the pipette tip up and down several times.
- Once the pellet has dissolved, place the sample at 55 °C for 10 minutes.
- Then, measure the concentration of each RNA sample using a Nanodrop spectrophotometer (2 μL of the RNA sample).
- Store the RNA samples at -80°C until further use.
III. First-Strand cDNA Synthesis (As per manufacturer s protocol using SuperScript first strand Synthesis kit by Invitrogen.)
- For each RNA sample add the following in a PCR tube:
RNA x μl 10mM dNTP mix M 1μl Oligo(dt) (0.5 μg/ μl) 1 μl DEPC treated water (8-x) μl Total Volume: 10 μl Note: x is the volume of RNA used for cDNA synthesis. Depending on the concentration of the RNA, 2-3 μg of RNA will be used for each reaction and the total volume of each reaction should be 10 μl. - Place the above reaction mix in a thermocycler at 65°C for 4 minutes.
- Remove the tubes from the theromocyler and place them on ice immediately for at least 2 minutes.
- Prepare the first-strand master mix by adding the following:
10X RT Buffer 2μl 25mM MgCl2 4 μl 0.1M DTT 2 μl RNase out 1 μl - Add 9 μl of master mix to each tube containing the RNA sample, mix by pipeting and centrifuge the mixture briefly.
- Place the tubes back in the thermocycler and incubate for 2 minutes at 42°C.
- Pause the thermocycler and add 1 μl of Superscript II reverse transcriptase enzyme (Invitrogen) to each tube. This step should be done quickly.
- Incubate the samples at 42°C for 1.5 hours.
- Terminate the reaction at 70°C for 15 minutes.
- Remove the tubes from the thermocycler and place them on ice.
- Add 1 μl of RNase H to each tube and place them back in the thermocycler at 37°C for 20 minutes.
- After taking the samples out of the thermocycler, add 20 μl of nuclease-free water in each tube. The volume of each sample is doubled at this stage.
- Check the concentration of each sample by using 2 μl of the sample. Take aliquot of each sample, to make the concentration of 20ng/μl for use in qRT-PCR. The amount of water to make up the samples to a concentration of 20ng/μl will depend on the initial concentration of the cDNA synthesized.
IV. qRT-PCR:
Primer Design and Determination of reference gene
- Design primers with a melting temperature (Tm) of 60°C and a product size of ~100bp.
- AP-PERI1 is used as the gene of interest for this experiment. A reference gene or internal control is needed for later analysis and normalization of the data. Ribosomal protein (AP-RP1) is used as the reference gene for this experiment.
- Prepare 5uM working stocks of the primers that will be used, including your reference gene.
Preparation of standards
- In order to calculate the efficiency of the primers being used standards should be made.
- Make a mix of the cDNA from the different samples that will be tested.
- Make 5X serial dilutions for each desired point in the graph. Four dilutions should be enough but five are highly recommended.
- The volume will vary depending on how many points and how many technical replicates are being used. There should be enough to make a standard for each gene being tested including the reference gene.
Plate Template
- Design a template indicating the sample name for each well in the qRT-PCR plate. Remember to include standards for each gene and at least two technical replicates per sample.
Set up the cycling parameters
- Turn the qRT-PCR machine on and enter the cycling parameters. The cycling parameters for this experiment is as follows:
Step 1:- 1 cycle: 95°C 3 min
- 40 cycles: 95°C 15 sec followed by 60°C 30 sec
(Optional: for determining the melting curve include the following steps)
- 1 cycle: 95°C 1 min
- 1cycle: 55°C 1 min
- 81 cycles: 55°C 30 sec
Plate set up for CFX96 (BioRad) machine
- Prepare the master mix for each gene (including the reference gene) in separate tube. For each well, the master mix consists of
Nuclease free water 2μl 2.5 X SYBR green 4 μl Primer forward 1 μl Primer reverse 1 μl Total volume 8 μl - While preparing the master mix include 1-2 extra reactions to account for pipetting errors.
- According to the previously designed template, add 2 μL (20 ng/ μL) of cDNA to each well. Then, add 8 μL of the master mix to each well, resulting in a total volume of 10 μL per reaction.
- Pipette up and down (2-3X) to make sure the sample is well mixed. Check that no liquid stays on the tip. To avoid cross contamination, use a new tip for each sample.
- Once the entire plate is setup, cover the plate with optical tape. Avoid touching the tape as the presence of grease can affect the reading. Then, centrifuge the plate at 500 rpm for 1 minute to ensure that all the products in the wells are at the bottom of the plate.
- Following centrifugation, check that there is no ice or water on the bottom of the plate. Finally, place the plate in the qRT-PCR machine and run the PCR program.
V. Diagrammatic Representation of the Experiment

Figure 1: Flow chart depicting the sequential order of steps for the gene expression study.
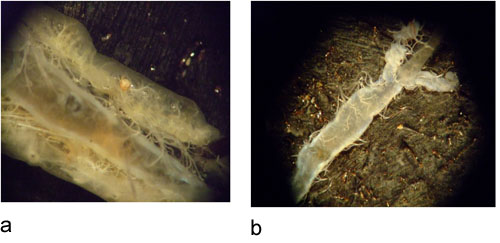
Figure 2: A) larval EAB dissection showing midgut in the middle. B) isolated midgut of larval EAB

Figure 3: A) Mean relative expression values (REVs) for a peritrophin gene (AP-PERI1) in different larval tissues including cuticle (Cu), midgut (MG) and fat bodies (FB). An EAB specific ribosomal protein (herein named, AP-RP1) was used as the internal control for normalizing the data obtained for the gene of interest, AP-PERI1. B) Relative fold change of AP-PERI1 in larval tissues. The cuticle tissue showed the least expression and therefore was taken as the calibrator (1X) sample (Pfaffl, 2001).

Figure 4: A) Mean relative expression values (REVs) for a peritrophin gene (AP-PERI1) in different developmental stages of EAB including larval instars (1st, 2nd, 3rd and 4th), prepupae (PP) and adults (A). An EAB specific ribosomal protein (AP-RP1) was used as the internal control for normalizing the data obtained for the gene of interest, AP-PERI1. B) Relative fold change of AP-PERI1 in various developmental stages. The PP sample showed the least level of mRNA levels and therefore was taken as the calibrator (1X sample).
VI. Conclusion
Quantitative real-time PCR (qRT-PCR) is an effective tool to diagnose mRNA levels in different insect tissues and developmental stages. Further, qRT-PCR has mostly been the key tool to validate data generated from high throughput gene expression analyses such as microarrays and the new-generation RNA-Seq.
Discussion
The threshold cycles (Ct) value is obtained for each sample in the qRT-PCR plate. For making the standard curve, the Ct value obtained for each dilution was plotted against the log of its concentration. The Ct values for the experimental samples were then plotted onto this dilution series standard curve. Target quantities were calculated from separate standard curves generated for each experiment i.e. tissue and developmental expression. The relative expression values (REVs) were then determined by dividing the quantitie...
Disclosures
No conflicts of interest declared.
Acknowledgements
We acknowledge the help provided by Lourdes Delta Arrueta Antequera (Department of Entomology, Ohio State University/OARDC, Wooster, OH) in the setup of the experiments. We thank Dr. Therese Poland (USDA, Forest Services, NRS) for send EAB larval samples. Help provided by Dr. Luis A Canas and Nuris M Acosta with the microscope setup is appreciated. Funding for this project was provided by State and Federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University.
References
- Lehane, M. J. Peritrophic matrix structure and function. Annu Rev Entomol. 42, 525-550 (1997).
- Mittapalli, O., Sardesai, N., Shukle, R. H. cDNA cloning and transcriptional expression of a peritrophin-like gene in the Hessian fly, Mayetiola destructor [Say]. Arc of Insect Bio. 64, 19-29 (2007).
- Pauchet, Y., Wilkinson, P., van Munster, M., Augustin, S., Pauron, D., ffrench-Constant, R. H. Pyrosequencing of the midgut transcriptome of the poplar leaf beetle Chrysomela tremulae reveals new gene families in Coleoptera. Ins Bio and Mol. 39, 403-413 (2009).
- Pfaffl, W. M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 2002-2007 (2001).
- Tellam, R. L., Lehane, M. J., Billingsley, P. F. . Biology of the Insect Midgut. , 86-114 (1996).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved