A subscription to JoVE is required to view this content. Sign in or start your free trial.
Expansion Pathology: A Method for High-resolution Optical Imaging of Clinical Tissue Samples
In This Article
Overview
In this video, we describe a method called expansion pathology, a variant of expansion microscopy, wherein the clinical tissue specimens are chemically expanded using a polymer network for microscopic examination of nanoscale structures.
Protocol
1. In Situ Polymerization of Specimens
- Incubate the specimen in an anchoring solution.
- Prepare the anchoring solution (typically 250 µL is enough to cover the tissue section) by diluting the AcX stock solution in 1x PBS to a concentration of 0.03 mg/mL for samples fixed with non-aldehyde fixatives or 0.1 mg/mL for samples fixed with aldehyde fixatives, which have fewer free amines available to react with AcX.
- Place the slide in a 100 mm Petri dish and pipette the anchoring solution over the tissue. Incubate for at least 3 h at RT or overnight at 4 °C.
- Incubate the samples in gelling solution.
- Prepare at least 100-fold excess volume of gelling solution. Per 200 µL, combine the following, in order: 188 µL of monomer solution, 4 µL of 0.5% 4HT stock solution (1:50 dilution, final concentration: 0.01%), 4 µL of 10% TEMED stock solution (1:50 dilution, final concentration 0.2%), and 4 µL of 10% APS stock solution (1:50 dilution, final concentration 0.2%).
NOTE: Gelling solution should be made immediately before use. The solution should be kept at 4 °C and the APS solution should be added last, to prevent premature gelling. - Remove the excess solution from the tissue section and place the slide in a 100 mm Petri dish. Add fresh, cold gelling solution to the sample and incubate the mixture on the tissue for 30 min at 4 °C, to allow diffusion of solution into the tissue.
- Prepare at least 100-fold excess volume of gelling solution. Per 200 µL, combine the following, in order: 188 µL of monomer solution, 4 µL of 0.5% 4HT stock solution (1:50 dilution, final concentration: 0.01%), 4 µL of 10% TEMED stock solution (1:50 dilution, final concentration 0.2%), and 4 µL of 10% APS stock solution (1:50 dilution, final concentration 0.2%).
- Construct a chamber on the slide around the sample (Figure 1A) without disturbing the gelling solution.
- Make spacers for the gelling chamber by thinly cutting pieces of cover glass using a diamond knife.
NOTE: To facilitate imaging post-expansion, the spacers should be close in thickness to the tissue specimen to reduce the amount of blank gel above the tissue. Number 1.5 glass can be used for standard clinical samples (5−10 µm). Cover glass pieces can be stacked for thicker samples. - Secure the spacers on either side of the tissue using droplets of water (~10 µL).
- Carefully place a cover glass lid over the slide, making sure to avoid trapping air bubbles over the tissue (Figure 1B).
- Make spacers for the gelling chamber by thinly cutting pieces of cover glass using a diamond knife.
- Incubate the sample at 37 °C in a humidified environment (such as a closed Petri dish with a damp wipe) for 2 h.
NOTE: The protocol can be paused here. The slide chamber can be stored inside a sealed Petri dish at 4 °C.
2. Sample Digestion
- Remove the lid of the gelling chamber by gently sliding a razor blade under the coverslip and slowly lifting the coverslip off the gel surface. Trim the blank gel around the tissue to minimize volume. Cut the gel asymmetrically to track the orientation of the gel after homogenization, since the sample will become transparent.
- Dilute ProK by 1:200 in digestion buffer (final concentration 4 U/mL) before use. Prepare enough solution to completely submerge the gel; a single well of a four-well plastic cell culture plate requires at least 3 mL per well.
- Incubate the sample in a closed container containing the digestion buffer for 3 h at 60 °C. If the sample does not detach from the slide during digestion, use a razor blade to gently remove the sample.
NOTE: The specimen should be completely submerged in digestion buffer to prevent the sample from drying out and placed in a covered container (small slide box, plastic well, Petri dish, etc.) that can be sealed with film.
3. Sample Expansion and Imaging
- Use a soft paintbrush to transfer the specimen into 1x PBS in a container compatible with the desired imaging system and large enough to accommodate the fully expanded gel. Make sure that the tissue is placed with the sample-side down if imaging on an inverted system or up if imaging on an upright system to minimize the distance from the imaging objective to the sample. Flip the gel using a soft paintbrush if needed.
NOTE: Side-illumination from an LED can be used to make them visible in liquid. A standard 6-well plate can accommodate samples that have a pre-expanded diameter less than 0.6 cm. A glass-bottom well plate should be used for imaging on an inverted system. - Wash the samples in 1x PBS at RT for 10 min. If desired, re-stain the sample with 300 nM DAPI as the digestion process washes away the DAPI stain. Remove PBS and stain with 300 nM DAPI diluted in 1x PBS for 20 min at RT, followed by a 10 min wash with 1x PBS at RT.
NOTE: The samples can be covered with 1x PBS and stored at 4 °C before proceeding to the next step. - To expand the samples, replace the PBS and wash with an excess volume of ddH2O (at least 10x the final gel volume) 3−5 times for 10 min each, at RT.
NOTE: After the 3rd or 4th wash, the specimen's expansion should begin to plateau. For storage, to prevent bacterial growth, the ddH2O can be supplemented with 0.002%−0.01% sodium azide (NaN3). In this case, the final expansion factor is reversibly reduced by 10%. - Perform fluorescence imaging using a conventional wide-field microscope, confocal microscope, or other imaging system of choice.
NOTE: To prevent gels from drifting, excess liquid can be removed from the well. Gels can also be immobilized with 1.5−2% low-melt agarose. Prepare 1.5−2%(w/v) low-melt agarose in water in a container 2−4 times the volume of solution. Warm the solution in a 40 °C water bath or in a microwave for 10−20 s to melt the solution. Pipette the melted agarose around the edges of the gel. After allowing the agarose to harden at RT or 4 °C, add water to the sample to prevent dehydration.
Results
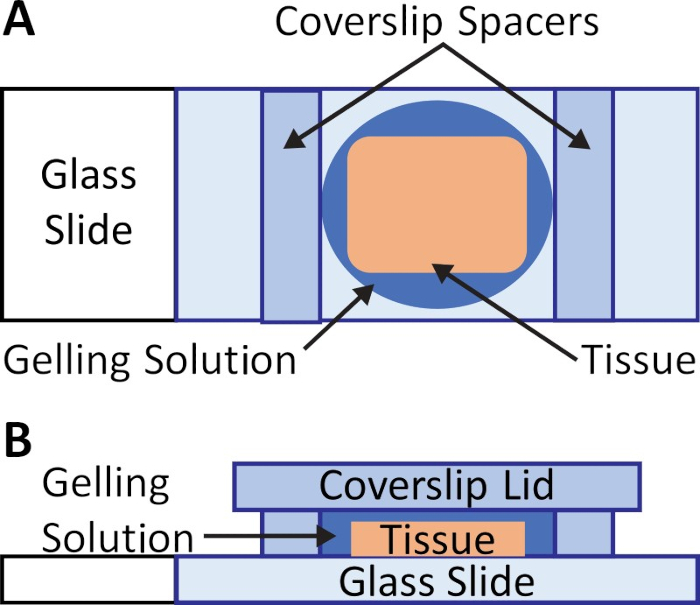
Figure 1: Gelation chamber for pathology samples. (A) Two spacers, such as two pieces of #1.5 cover glass cut with a diamond knife, are placed on either side of the tissue after incubating the sample in gelling solution at 4 °C. The spacers should be thicker than the tissue slices, to prevent compression of the sample. (B) A lid, such a...
Disclosures
Materials
| Name | Company | Catalog Number | Comments |
| 6-well glass-bottom plate (#1.5 coverglass) | Cellvis | P06-1.5H-N | |
| Acetone | Fischer Scientifc | A18-500 | |
| Acrylamide | Sigma Aldrich | A8887 | |
| Acryloyl-X SE, (AcX) | Invitrogen | A20770 | |
| Agarose | Fischer Scientifc | BP160-100 | |
| Ammonium persulfate (APS) | Sigma Aldrich | A3678 | Initiatior |
| DAPI (1 mg/mL) | Thermo Scientific | 62248 | Nuclear stain |
| Diamond knife No. 88 CM | General Tools | 31116 | |
| Ethanol | Pharmco | 111000200 | |
| Ethylenediaminetetraacetic acid (EDTA) 0.5 M | VWR | BDH7830-1 | |
| FFPE Kidney Sample | USBiomax | HuFPT072 | |
| Forceps | |||
| Micro cover Glass #1 (24 mm x 60 mm) | VWR | 48393 106 | |
| Micro cover Glass #1.5 (24 mm x 60 mm) | VWR | 48393 251 | |
| N,N,N′,N′-Tetramethylethylenediamine (TEMED) | Sigma Aldrich | T9281 | Accelerator |
| N,N′-Methylenebisacrylamide | Sigma Aldrich | M7279 | |
| Nunclon 6-Well Cell Culture Dish | Thermo Fisher | 140675 | |
| Nunc 15 mL Conical | Thermo Fisher | 339651 | |
| Nunc 50 mL Conical | Thermo Fisher | 339653 | |
| Orbital Shaker | |||
| Paint brush | |||
| pH Meter | |||
| Phosphate Buffered Saline (PBS) 10x Solution | Fischer Scientifc | BP399-1 | |
| Plastic Petri Dish (100 mm) | Fischer Scientifc | FB0875713 | |
| Proteinase K (Molecular Biology Grade) | Thermo Scientific | EO0491 | |
| Razor blade | Fischer Scientifc | 12640 | |
| Safelock Microcentrifuge Tubes 1.5 mL | Thermo Fisher | 3457 | |
| Safelock Microcentrifuge Tubes 2.0 mL | Thermo Fisher | 3459 | |
| Sodium acrylate | Sigma Aldrich | 408220 | |
| Sodium chloride | Sigma Aldrich | S6191 | |
| Sodium citrate tribasic dihydrate | Sigma Aldrich | C8532-1KG | |
| Tris Base | Fischer Scientifc | BP152-1 | |
| Triton X-100 | Sigma Aldrich | T8787 | |
| Xylene | Sigma Aldrich | 214736 |
This article has been published
Video Coming Soon
Source: Klimas, A. et al. Nanoscopic Imaging of Human Tissue Sections via Physical and Isotropic Expansion. J. Vis. Exp. (2019)
Copyright © 2025 MyJoVE Corporation. All rights reserved