A subscription to JoVE is required to view this content. Sign in or start your free trial.
Electrophoretic Mobility Shift Assay for Detection of Transcription Factor-DNA Complexes
In This Article
Overview
In this video, we demonstrate the electrophoretic mobility shift assay to detect protein-DNA interactions using infrared fluorescent-labeled DNA probes. When the protein binds to the DNA probe, it forms a large complex that migrates slowly on and appears as a shifted band on the gel.
Protocol
1. Binding Reaction and Electrophoresis
- Prepare 1 mL of 5x binding buffer by mixing 50 µL of 1 M Tris-HCl, pH 7.5; 10 µL of 5 M NaCl; 200 µL of 1 M KCl, 5 µL of 1 M MgCl2, 10 µL of 0.5 M EDTA, pH 8.0; 5 µL of 1 M DTT; 25 µL of 10 mg/mL BSA, and 695 µL of ddH2O.
NOTE: The 5x Binding Buffer can be aliquoted and stored at -20 °C. Another point to consider is that different transcription factors will have different modifications to the binding buffer. - Right before setting up binding reactions, pre-run the 5% native polyacrylamide gel in 0.5x TBE + 2.5% glycerol to remove all traces of ammonium persulfate at 80 V for 30 - 60 min, or until the current no longer varies with time.
- Set up the binding reaction in the final volume of 20 µL.
- Mix 4 µL of 5x binding buffer, 80 - 200 ng purified protein A (in 50% glycerol, i.e., SOX-2), 1 µL of 0.1 µM Dye-conjugated probe (final concentration: 5 nM), and ddH2O.
- Optional: When cold probe competitors are required, add various concentrations (2x, 10x, 25x, 50x, 100x, etc.) of cold probes.
- Optional: When the interaction between proteins A and B (i.e., SOX-2 and LIM-4) is tested, add 80 - 200 ng purified protein B (in 50% glycerol, i.e., LIM-4).
- Optional: When nuclear extract preparation is used and the specific binding of protein A is to be validated, add an antibody specific to protein A (i.e., anti-6xHis, anti-FLAG, etc.).
- Incubate at R/T in the dark for 15 min.
- Load all of the binding reactions onto the gel and run the gel at 10 V/cm to the desired distance.
2. Imaging
NOTE: The gel was scanned directly in the glass plates with an advanced infrared imaging system. Therefore, the gel can be resolved further and scanned repeatedly (Figures 1C and 2). A near-infrared fluorescent imaging system primarily for Western blots was also tested to scan the gel, but only the advanced infrared imaging system was able to scan the gel with good resolution. The methodology described is specific to a particular infrared imaging software, although other software packages may be used.
- Clean the bed of the scanner with ddH2O and dry well before scanning. Wipe dry the glass plates of the gel and place the plates containing the gel on the scanner bed.
- Open the infrared imaging software and go to the 'Acquire' tab. When the thinner plate (1 mm) is placed on the scanner bed, use the settings of Channel: 700, Intensity: Auto, Resolution: 169 µm, Quality: medium, and Focus offset: 1.5 mm. When the thicker plate (3 mm) is placed on the scanner bed, use the settings of Channel: 700, Intensity: Auto, Resolution: 84 µm, Quality: medium, and Focus offset: 3.5 mm. Focus offset depends on the thickness of the glass plate.
- Select the area that the gel occupies on the scanner. Click 'Start' to begin the scan.
Results
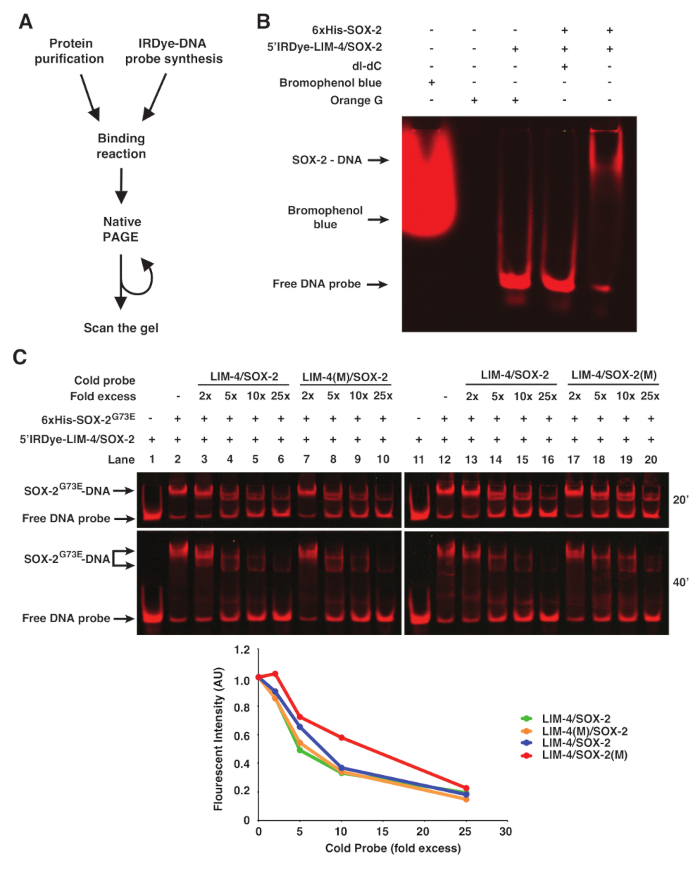
Figure 1. fEMSA using Infrared Fluorescent Dye-labeled Oligonucleotides. (A) Flow chart of fEMSA. (B) The effect of bromophenol blue dye, orange G, and dI-dC (1 µg) on fEMSA. 5 nM of 5'Dye-LIM-4/SOX-2 probe was used.(C)fEMSA showing binding specificity of 6xHis-SOX-2G73E to the SOX-2 target site of the LIM-4/SOX-2 element. 5 nM of 5'Dye-LIM-4/SOX-2 probe w...
Disclosures
Materials
| Name | Company | Catalog Number | Comments |
| 30% Acrylamide/Bis Solution, 37.5:1 | Bio-Rad | 1610158 | Acrylamide is harmful and toxic. |
| Bovine Serum Albumin molecular-biology-grade | New England Biolabs | B9000S | |
| 5'IRDye700-labeled DNA Oligos | Integrated DNA Technologies | Custom DNA oligo | These are referred as 5'Dye-labeled or infrared fluorescent dye-labeled DNA oligos in the manuscript. The company will custom-synthesize 5' IRDye-labeled DNA oligonucleotides. Requires minimum 100μM scale synthesis and HPLC purification. |
| Mini-PROTEAN Vertical Electrophoresis Cell | Bio-Rad | 1658000FC | Device |
| Odyssey CLx Infrared Imaging System | LI-COR Biotechnology | Odyssey CLx Infrared Imagng System | Device |
| Odyssey Fc Imaging System | LI-COR Biotechnology | Odyssey Fc Dual-Mode Imaging System | Device |
| Image Studio software (version 4.0) | LI-COR Biotechnology | Image Studio software | Software |
| Orange G | Sigma-Aldrich | O3756-25G | Chemical |
| 6x Orange loading dye | 0.25% Orange G; 30% Glycerol | ||
| 0.5x TBE | 45 mM Tris-Borate; 1 mM EDTA | ||
| 1x TE | 10 mM Tris-HCl; 1 mM EDTA, pH8.0 | ||
| 1x STE | 100 mM NaCl; 10 mM Tris-HCl, pH8.0; 1 mM EDTA | ||
| 5x Binding Buffer | 50 mM Tris-HCl, pH7.5; 50 mM NaCl; 200 mM KCl; 5 mM MgCl2; 5 mM EDTA, pH8.0; 5 mM DTT; 250 ug/ml BSA |
This article has been published
Video Coming Soon
Source: Hsieh, Y. W. et al., An Optimized Protocol for Electrophoretic Mobility Shift Assay Using Infrared Fluorescent Dye-labeled Oligonucleotides. J. Vis. Exp. (2016)
Copyright © 2025 MyJoVE Corporation. All rights reserved