A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
High-throughput Parallel Sequencing to Measure Fitness of Leptospira interrogans Transposon Insertion Mutants During Golden Syrian Hamster Infection
In This Article
Summary
We describe here a technique that combines transposon mutagenesis with high-throughput sequencing to identify and quantify transposon leptospiral mutants in tissues after a challenge of hamsters. This protocol can be used to screen mutants for survival and dissemination in animals and can also be applied to in vitro studies.
Abstract
In this manuscript, we describe a transposon sequencing (Tn-Seq) technique to identify and quantify Leptospira interrogans mutants altered in fitness during infection of Golden Syrian hamsters. Tn-Seq combines random transposon mutagenesis with the power of high-throughput sequencing technology. Animals are challenged with a pool of transposon mutants (input pool), followed by harvesting of blood and tissues a few days later to identify and quantify the number of mutants in each organ (output pools). The output pools are compared to the input pool to evaluate the in vivo fitness of each mutant. This approach enables screening of a large pool of mutants in a limited number of animals. With minor modifications, this protocol can be performed with any animal model of leptospirosis, reservoir host models such as rats and acute infection models such as hamsters, as well as in vitro studies. Tn-Seq provides a powerful tool to screen for mutants with in vivo and in vitro fitness defects.
Introduction
Identification of virulence genes for some bacteria, such as Leptospira spp., is difficult because of the limited number of genetic tools available. One commonly used approach is the creation of a collection of mutants by random transposon mutagenesis followed by the identification of the insertion site in each mutant and virulence testing of individual transposon mutants in an animal model. This approach is time-consuming, expensive, and requires a large number of animals.
When random mutagenesis was first developed for the pathogen Leptospira interrogans, genes involved in virulence were identified by testing individual mutants in an animal model1. Mutants were selected based on criteria such as their potential roles in signaling or motility or their predicted outer membrane or surface location. As the majority of leptospiral genes encode hypothetical proteins of unknown function2, selecting mutants based on these criteria limits the ability to discover novel leptospiral virulence genes.
More recently, pools of L. interrogans transposon mutants were screened for infectivity in the hamster and mouse models3. Each animal was challenged with a pool of up to 10 mutants. Infectivity of a mutant was scored as positive if it was detected by PCR of cultures obtained from blood and kidney. PCR testing was laborious because it required an individual PCR reaction for each mutant in the pool. Because the frequency of each mutant in the cultures was not quantified, the approach was biased towards identification of highly attenuated mutants.
We describe a transposon sequencing (Tn-Seq) technique, as a strategy to more efficiently screen for virulence genes. Tn-seq consists of the creation of a library of mutants by transposon mutagenesis followed by massive parallel sequencing4,5,6. Briefly, transposon mutants are pooled, inoculated into animals, and later recovered from different organs (output pools). The DNA from the output pools is extracted and digested with restriction enzymes or sheared by sonication. Two rounds of PCR targeting the junctions of the transposon insertion sites are performed. This step enables the addition of the adaptors necessary for the sequencing. The resulting PCR products are analyzed by high-throughput sequencing to identify the transposon insertion site of each mutant of the pool along with their relative abundance, which is compared to the initial composition of the pool of mutant.
The primary advantage of this approach is the ability to simultaneously screen a large number of mutants with a small number of animals. Tn-Seq does not require the prior knowledge of the transposon insertion sites which increases the chances of discovering new Leptospira-specific genes involved in virulence with less time and greater efficiency. Because leptospiral burden in tissues is relatively high in rodent models susceptible to lethal infection (typically 104 to 108 bacteria/g of tissue)7,8,9 as well as in reservoir hosts10,11, tissues can be directly analyzed without the need to culture, reducing biases due to in vitro growth.
In Tn-Seq studies with most bacterial pathogens described to date, the high frequency of insertional mutagenesis allowed infection with large pools containing mutants collectively having multiple closely-spaced transposon insertions within every gene4,12,13,14. Tn-Seq has also been developed for a bacterium for which the mutagenesis frequency is much lower6. With Leptospira, a library of transposon mutants can be generated by introducing the transposon on a mobilizable plasmid by conjugation as described by Slamti et al15. However, the frequency of transposon mutagenesis of L. interrogans is low. When the Himar1 transposon was introduced on a conjugative plasmid, the transconjugant frequency was reported to be only 8.5 x 10-8 per recipient cell with the Lai strain of L. interrogans16 and is likely to be similarly poor with most other strains of L. interrogans. The protocol described here is in part based on that developed for Borrelia burgdorferi, in which the frequency of transposon insertional mutagenesis is also low6.
For our pilot experiment with the protocol17, we conducted transposon mutagenesis with L. interrogans serovar Manilae strain L495 because of the success of other groups in isolating transposon insertion mutants in the strain along with its low LD50 (lethal dose) for virulence1. We screened 42 mutants by Tn-Seq and identified several mutant candidates defective in virulence, including two with insertions in a candidate adenylate cyclase gene. Individual testing of the two mutants in hamsters confirmed that they were deficient in virulence17.
Protocol
CAUTION: Pathogenic strains of Leptospira spp. must be handled under Biosafety Level 2 (BSL-2) containment procedures. Appropriate personal protective equipment (PPE) must be worn. A Class II biosafety cabinet must be used for all manipulations of pathogenic Leptospira spp.
1. Creation of the Transposon Mutant Library15
- Transfer of the transposon into Leptospira spp. by conjugation (Figure 1)
- Inoculate a volume of exponential-phase culture of pathogenic Leptospira spp. corresponding to 107 cells into 10 mL of Ellinghausen-McCullough-Johnson-Harris medium (EMJH)18,19. Incubate at 30 °C with 150 rpm shaking until the density reaches 2 - 8 x 108 cells/mL.
NOTE: The doubling time of pathogenic leptospires is 12 to 24 h, depending on the strain. - Inoculate 50 µL of the donor Escherichia coli strain β216320 carrying the mobilizable transposon plasmid (pCjTKS2)16 into 5 mL of Luria Broth (LB) supplemented with 0.3 mM of 2,6-diaminopimelicacid (DAP), 50 µg/mL of kanamycin (Km), and 50 µg/mL of spectinomycin (Spc) and place overnight in a 37 °C incubator at 255 rpm.
- Inoculate 60 µL of E. coli cells into 3 mL of EMJH supplemented with 0.3 mM of DAP (EMJH+DAP). Incubate at 37 °C at 255 rpm agitation for 3 - 4 h up to an OD600nm≈ 0.3.
- To assemble the filtration unit (Figure 1), place the base on a 125 mL side-arm Erlenmeyer flask, position an acetate-cellulose filter (pore size 0.1 mm; diameter 25 mm) on the base, and clamp the funnel onto the base. Connect the filtration unit to a vacuum system.
- Add 5 mL of Leptospira spp. culture and 0.5 mL of E. coli culture into the funnel.Vacuum the liquid through the filter.
- Transfer the filter with the surface of bacteria facing up onto an EMJH+DAPplate. Incubate at 30 °C overnight with the filter facing up.
- Place the filter into a 15 mL tube containing 1 mL of EMJH and vortex for 10 s to release the bacteria into the media. Spread 200 µL of the suspension onto 5 EMJH platescontaining 50 µg/mL of Km using 10-15 1 mm sterile glass beads or a sterile disposable spreader. Wrap the plates with parafilm and incubate them upside down at 30 °C for 3 to 4 weeks until colonies are visible.
- Transfer colonies individually into 3 mL of EMJH containing 50 µg/mL of Km (EMJH+Km) at 30°C under 150 rpm agitation for 7 to 10 days until the culture reaches a density of ≈ 108/mL.
NOTE: Cultures can be stored at -80 °C or in liquid nitrogen (with 4% glycerol).
- Inoculate a volume of exponential-phase culture of pathogenic Leptospira spp. corresponding to 107 cells into 10 mL of Ellinghausen-McCullough-Johnson-Harris medium (EMJH)18,19. Incubate at 30 °C with 150 rpm shaking until the density reaches 2 - 8 x 108 cells/mL.
- Identification of the transposon insertion site by nested PCR (Figure 2)
- Lyse 50 µL of each transposon mutant in PCR tubes by incubation at 95 °C for 15 min.
NOTE: DNA can be purified instead, using a DNA extraction kit. - Prepare the PCR mix with Deg1 and Tnk1 primers (Table 3) according to Table 1. Transfer 23.7 µL of the mix to each PCR tube and add 1.3 µL of lysed cells. Run the program: 95°C for 5 min; 40 cycles: 95 °C for 15 s, 40 °C for 1 min, 72 °C for 2 min; 72 °C for 10 min.
- Make the PCR mix with Tag and TnkN1 primers (Table 3) according to Table 2. Transfer 24.2 µL of the mix to each PCR tube and add 0.8 µL of PCR #1 reaction. Run the program: 95 °C for 5 min; 35 cycles: 95 °C for 15 s, 55 °C for 30 s, 72 °C for 2 min; 72 °C for 10 min.
- Run 3 µL of PCR products on a 1% agarose gel with 1X Tris-Acetate-EDTA (TAE) buffer at 10-15 V/cm (Figure 2B).
- Purify PCR products of positive samples using a PCR purification kit. Elute DNA with the lowest volume allowed by the kit to maximize the concentration of DNA recovered.
- Send purified PCR products for Sanger sequencing using the TnkN1 primer (Table 3).
- Identify the insertion sites by comparing the resulting sequence with the parental strain's genome sequence by BLASTN analysis (http://blast.ncbi.nlm.nih.gov/) or using the SpiroScope database (http://www.genoscope.cns.fr/agc/mage)21.
- Confirm the insertion site of the transposon by PCR using primers annealing to the flanking host sequences.
NOTE: The transposon increases the size of the wild-type sequence by ≈ 2 kb.
- Lyse 50 µL of each transposon mutant in PCR tubes by incubation at 95 °C for 15 min.
2. Animal Experiment (Figure 3)
- Culture of Leptospira mutants
- Grow individually each selected transposon mutant in 10 mL of EMJH+Km at 30 °C at 150 rpm agitation to a density of 107-108 leptospires/mL.
- Count the leptospires by dark-field microscopy with a Petroff-Hausser counter or as described by Miller23.
- Dilute each culture in EMJH to the same density, for instance 106 cells/mL.
- To assemble the input pool, mix together the diluted cultures in equal volumes.
NOTE: Include controls in the input pool by adding mutants with known fitness defects such as loa2217,24 and mutants with unaltered fitness such as ligB17,25, respectively. Pools of mutants can be stored with 4% glycerol at -80 °C or in liquid nitrogen.
- Challenge
NOTE: The methods described here were approved by the Veterans Affairs Greater Los Angeles Institutional Animal Care and Use Committee (protocol #09018-14).- Inject intraperitoneally 1 mL of the input pool to each animal with an insulin syringe U-100 with 26G x ½" needle.
NOTE: In the pilot experiment, 8 animals were challenged with 1 mL of the input pool, i.e., 106 bacteria total17. The infection was allowed to proceed for 4 days prior to euthanasia. - Collect 10 mL of the input pool, and spin for 20 min at 3,220 x g. Carefully remove the supernatant without disturbing the pellet. Store the cell pellet at -80˚C until use (step 3.1.3).
- Monitor the animals daily until they are terminated at the pre-determined endpoint. Weigh the hamsters daily and look for endpoint criteria: loss of appetite, gait or breathing difficulty, prostration, ruffled fur, or loss of > 10% of the maximum weight attained.
- Inject intraperitoneally 1 mL of the input pool to each animal with an insulin syringe U-100 with 26G x ½" needle.
- In vitro experiment
- On the day of challenge, inoculate 3 flasks of 25 mL of EMJH+Km with 5 mL of the input pool. Grow cultures at 30 °C under 150 rpm agitation.
- Count the leptospires daily by using one of the methods of section 2.1.2. When the density reaches ≈ 1 x 108/mL, spin down each suspension for 20 min at 3,220 x g.
- Store the cell pellets at -80 °C until use.
- Harvesting and storage of the tissues
- Euthanize the animals by isoflurane inhalation followed by bilateral thoracotomy26.
- Immediately collect 1 to 2 mL of blood by cardiac puncture with a 3 mL syringe and a 25G x 5/8" needle. Transfer the blood into tube containing EDTA. Mix by inversion, 5 to 6 times.
- Collect one kidney and ⅓ to ½ of the ventral median lobe of the liver into cryotubes.
- Store tissues at -80 °C until use.
3. Construction of Genomic Libraries for High-throughput Sequencing (Figure 4)
- DNA extraction
- DNA extraction from blood.
- Transfer 100 µL of blood from the EDTA tube to a microcentrifuge tube.
- Purify DNA using a DNA extraction kit. Follow the manufacturer's instructions.
- DNA extraction from tissues.
- Using scalpels and scissors, dice between 50 to 80 mg of each organ into small pieces (1 mm x 1 mm), and transfer them into a sterile screw-cap dry tube. Measure the weight of tissue with a precision balance.
- Add 500 µL of sterile PBS into the tube.
- Homogenize samples by using the disruptor for 1 min at 5 movements per second.
- Calculate the volume corresponding to 25 mg of tissue using the following equation:
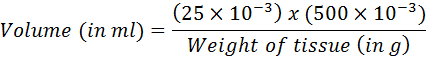
- Transfer the calculated volume into the DNA extraction kit. Proceed with DNA purification following the manufacturer's instructions.
- DNA extraction from the input pool and in vitro cultures.
- Thaw bacterial pellets at room temperature for 5-10 min.
- Proceed with DNA extraction following the instructions accompanying the kit.
- Store DNA at -80 °C until use.
- DNA extraction from blood.
- Shearing the DNA (Figure 5)
- Transfer 50 µL of extracted DNA onto a 1.5 mL microcentrifuge tube.
- Place the tubes into the rack of the sonicator cup horn filled with cold water (4 °C).
- Run the sonicator for 3 min at 80% intensity with 10 s on pulse and 5 s off pulse.
CAUTION: Wear ear muffs or ear plugs to protect hearing. - Run 2.5 µL of the sheared DNA on a 2% agarose gel to confirm that most of the DNA is <600 bp in size.
- Addition of the C-tail
- Measure DNA concentration with a small volume spectrophotometer.
- Calculate volume corresponding to 500 ng of DNA by using the following equation:
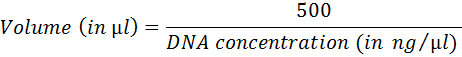
- Prepare tailing reaction (Table 4). Incubate 1 h at 37 °C; inactivate at 75 °C for 20 min.
NOTE: For samples that require adjustment to a volume greater than 14.5 µL, increase the final volume of reaction to 40 µL and scale up the remaining components accordingly. - Clean the samples with a PCR purification kit. Elute the DNA with 12 µL of elution buffer.
- Nested PCR
- PCR #1
- Prepare PCR mixes according to Tables 3 and 5. Transfer 22 µL of the library mix to each PCR tube and add 3 µL of purified DNA. Proceed similarly with the control mix.
NOTE: Primer TnkN317 is specific to the transposon and primer olj3766 is specific to the C-tail. The control mix is lacking the TnkN3 primer, which specifically targets the transposon. - Run the following program: 95 °C for 2 min; 24 cycles: 95 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min; 72 °C for 2 min.
- Prepare PCR mixes according to Tables 3 and 5. Transfer 22 µL of the library mix to each PCR tube and add 3 µL of purified DNA. Proceed similarly with the control mix.
- PCR #2.
- Prepare the second PCR mixes according to Tables 3 and 6. Transfer 49 µL of the library mix to each PCR tube and add 1 µL of PCR #1 library reaction. Transfer 24.5 µL of the control mix to each PCR tube and add 0.5 µL of PCR #1 control reaction.
NOTE: Primer pMargent2 is specific to the transposon, and the IP primers6 contain six-base-pair barcode and specific sequences recognized by the next-generation sequencing platform. - Run the following program: 95 °C for 2 min; 18 cycles: 95 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min; 72 °C for 2 min.
- Prepare the second PCR mixes according to Tables 3 and 6. Transfer 49 µL of the library mix to each PCR tube and add 1 µL of PCR #1 library reaction. Transfer 24.5 µL of the control mix to each PCR tube and add 0.5 µL of PCR #1 control reaction.
- Run 3 µL on a 2% agarose gel. The library should show a smear with the majority of the signal between 200 to 600 bp (Figure 6) and no amplification for the control reaction.
- PCR #1
- Purification of PCR products.
NOTE: Clean the genomic libraries with a PCR purification kit following the manufacturer's instructions. Elute the DNA with 30 µL of elution buffer. - Measure the DNA concentration using a fluorometer. Average 2 to 3 reads.
- Calculate the volume of each library equivalent to 15 or 20 ng using the equation below:
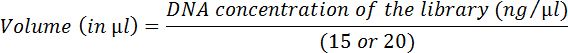
- Mix all libraries together according to the previous calculations. Determine the molar concentration of the DNA with the following website's equation:
http://www.molbiol.edu.ru/eng/scripts/01_07.html
CAUTION: DNA concentration and volume requirements depend on the sequencing platform.
4. High-throughput Sequencing and Data Analysis
- Sequencing
- Sequence libraries as 64 bp single-end reads using the custom sequencing primer pMargent3 and the standard commercial sequencing primer. The sequencing platform will provide you FastQ files with all sequencing reads.
- Analysis with Galaxy software
- Download the genome sequence file
- In the SpiroScope database homepage (see step 1.2.7 for link), select the organism used for the experiment and click "LOAD INTO GENOME BROWSER".
- In the toolbar (near the top of the homepage), select "Search/Export > Download data", in the "Sequence (fasta)" line, click "Genome" to download the sequence.
- Open the file with Notepad (PC) or TextEdit (Mac) and rename the chromosome (for example "ChrI"). Maintain the fasta format.
- Follow the same steps to download the sequence of chromosome II. Combine both chromosome sequences into a single .txt file by copying and pasting.
NOTE: Chromosome sequences can also be downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov) and combined into a single .txt file.
- Upload files onto the Galaxy server
NOTE: Galaxy is an open source, web-based platform for managing data intensive bioinformatics workflows27,28,29,30 and can be accessed at https://usegalaxy.org/.- In the Tools menu, select "Get data > Upload File from your computer". Drag and drop the .fastq file(s) generated by the sequencing platform into the window then click "Start".
- Follow the same steps to upload the Leptospira genome sequence .txt file.
- Groom reads
- Select "NGS: QC and manipulation > FASTQ Groomer" from the Tools menu.
- Next to File to groom, select the libraries uploaded in step 4.2.2. In Input FASTQ quality scores type, select the appropriate sequencing system. For Advanced Options, leave Hide Advanced Office selected.
- Click "Execute".
- Remove sequencing artifacts
- Select "NGS: QC and manipulation > Remove sequencing artifacts". Next to Library to filter, select the groomed files generated in step 4.2.3. Click "Execute".
- Remove C-tail sequences
NOTE: Repeat these steps once or twice to ensure all C-tails are removed.- Select "NGS: QC and manipulation > Clip adapter sequences".
- Select or enter the following:
Library to clip: select the files generated in step 4.2.4.
Minimum sequence length: 15
Source: enter custom sequence
Enter custom clipping sequence: CCCCCCC
Enter non-zero value to keep the adapter sequences and x bases that follow it: 0
Discard sequences with unknown (N) bases: Yes
Output options: Output both clipped and non-clipped sequences - Click "Execute".
- Remove adaptor sequences
- Select "NGS: QC and manipulation > Clip adapter sequences".
- Select or enter the following:
- Library to clip: select files generated in step 4.2.5.
- Minimum sequence length: 15
- Source: enter custom sequence
- Enter custom clipping sequence: CGTATGCCGTCTTCTGCTTG
- Enter non-zero value to keep the adapter sequences and x bases that follow it: 0
- Discard sequences with unknown (N) bases: yes
- Output options: Output both clipped and non-clipped sequences
- Click "Execute".
- Filter reads based on their quality
- Select "NGS: QC and manipulation > Filter by quality".
NOTE: This tool selects reads based on quality scores. - Select the following:
Library to filter: select the files generated in step 4.2.6.
Quality cut-off value: 20
Percent of bases in sequence that must have quality equal to/higher than cut-off value: 95 - Click "Execute".
NOTE: With these settings, reads shorter than 20 nucleotides or with a quality score of 20 or less for 95% of the cycles are discarded. Adapt stringency settings to your experiment.
- Select "NGS: QC and manipulation > Filter by quality".
- Map reads32
- Select "NGS: Mapping > Bowtie2"32.
- Select the following in the fields in the main window:
Is this single or paired library: single-end
FASTQ file: select the library filtered for quality from step 4.2.7.
Write unaligned (in fastq format) reads to separate file(s): no
Write aligned (in fastq format) reads to separate file(s): no
Will you select a reference genome from your history or use a built-in index?: Use a genome from the history and build index
Select the reference genome: select the Leptospira genome.txt file uploaded in step 4.2.2.
Set read groups information?: do not set
Select analysis mode: 1: default setting only
Do you want to use presets?: No, just use defaults
Save the bowtie2 mapping statistics to the history: No
Job resource parameters: Use default job resource parameters - Click "Execute" to align reads to the genome.
- Convert files
- Select "NGS: SAMtools > BAM-to-SAM convert BAM to SAM".
- Select the following:
BAM file to convert: Select mapped library from step 4.2.8.
Header options: include header in SAM output (-h) - Click "Execute".
- Convert files
- Select "NGS: SAMtools > Convert SAM to interval".
- Select the following:
Select dataset to convert: Select the SAM mapped library file generated in step 4.2.9.
Print all?: Yes - Click "Execute".
- Sort reads
- Select "Filter and Sort > Sort data in ascending or descending order".
- Select the following:
Sort dataset: Select the interval file generated in step 4.2.10.
on column: 2
with flavor: Numerical order
everything in: Ascending order - Click "Execute".
- Select reads matching chromosome I
- Select "Filter and Sort > Select lines that match an expression".
- Select the following:
Select lines from: select file with sorted reads generated in step 4.2.11.
that: Matching
the pattern: Enter the name of chromosome I as determined in step 4.2.1.3 (e.g., "ChrI"). - Click "Execute".
- Select reads matching chromosome II
Proceed following step 4.2.12. - Group reads according to the insertions sites in chromosome I
- Select "Join, Subtract and Group > Group data by a column and perform aggregate operation on other columns".
- Select the following:
Select data: select resulting file from step 4.2.12.
Group by column: 2
Ignore case while grouping?: No
Ignore lines beginning with these characters: Ø
Operation > + insert operation
Type: Count
On column: 2
Round result to nearest integer: NO - Click "Execute".
- Group reads according to the insertion sites in chromosome II
Proceed following step 4.2.14. - Sort insertion sites on chromosome I
- Select "Filter and Sort > Sort data in ascending or descending order".
- Select the following:
Sort Dataset: Select file from step 4.2.14.
on column: 1
with flavor: Numerical order
everything in: Ascending order - Click "Execute".
- Sort insertion sites on chromosome II
Proceed following step 4.2.16.
- Download the genome sequence file
- Statistical analysis
- Transfer the Galaxy data into spreadsheet files by copying and pasting the two columns from steps 4.2.16. and 4.2.17. into an Excel file.
NOTE: The first column is the nucleotide coordinate of the transposon insertion site, and the second column is the number of reads at each insertion site. - Identify the gene harboring the transposon using the nucleotide coordinate provided in the table.
NOTE: For example, a transposon inserted at nucleotide #30718 of chromosome I is located in the LIC10024 gene, which spans nucleotides 29263-31539 in chromosome I. - Calculate relative frequencies (F) for each mutant in each tissue and in the input pool following the equation below:
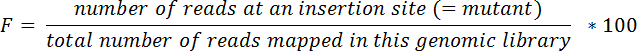
- Calculate output/input ratios (R) for each mutant and tissue using the equation below:
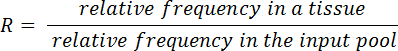
- Wilcoxon signed-rank test
- Normalize all output/input ratios by setting the median ratio for each tissue in each animal to 1.0. A ratio of 1.0 is neutral, >1.0 is disadvantageous, and <1.0 is advantageous33.
- Compare output/input ratios to 1.0 (neutral fitness) by using the Wilcoxon rank test with P values <0.05 considered statistically significant.
- Transfer the Galaxy data into spreadsheet files by copying and pasting the two columns from steps 4.2.16. and 4.2.17. into an Excel file.
Results
Creation of a library of transposon mutants in L. interrogans by conjugation requires a filtration unit, as shown in Figure 1. We recovered 100-200 transconjugants from each mating.
The transposon insertion site is identified in each mutant by sequencing the PCR product generated by semi-random PCR that targets the end of the transposon and adjacent host sequences15 ...
Discussion
Although results from our pilot experiment for hamster challenged intraperitoneally with 42 L. interrogans mutants are presented17, we expect that larger pools of mutants can be screened by Tn-Seq. Because the frequency of transconjugants is low (100-200 transconjugants/mating), several matings are necessary to generate a sufficient number of mutants for large Tn-Seq experiments. Maintaining a large number of mutants in liquid cultures presents logistical challenges that must be addressed...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a Veterans Affairs Merit Award (to D.A.H.) and a National Institute of Health grant R01 AI 034431 (to D.A.H.).
Materials
| Name | Company | Catalog Number | Comments |
| Kanamycin sulfate from Streptomyces kanamyceticus | Sigma-Aldrich | K4000 | |
| 2,6-diaminopimelic acid | Sigma-Aldrich | D1377 | |
| Spectinomycin dihydrochloride pentahydrate | Sigma-Aldrich | S4014 | |
| Axio Lab A1 microscope with a darkfieldcondenser | Zeiss | 490950-001-000 | |
| DNeasy blood and tissue kit | Qiagen | 69504/69506 | |
| MinElute PCR Purification | Qiagen | 28004/28006 | |
| QIAquick PCR purification kit | Qiagen | 28104/28106 | |
| Model 505 Sonic Dismembrator | Fisher Scientific | FB-505 | |
| 2.5" Cup horn | Fisher Scientific | FB-4625 | |
| Bead Ruptor 24 | Omni International | 19-010 | Step 3.1.2.4 |
| Terminal deoxynucleotidyl transferase | Promega | M828C | |
| Master mix Phusion | Thermo Scientific | F531 | Preparation of genomic libraries, step 3.4. |
| DreamTaq Master Mix | Thermo Scientific | K9011/K9012 | Identification of the transposon insertion site, step 1.2. |
| dCTP | Thermo Scientific | R0151 | |
| ddCTP | Affymetrix/ USBProducts | 77112 | |
| T100 Thermal cycler | BioRad | 1861096 | |
| Qubit 2.0 fluorometer | Invitrogen | Q32866 | step 3.6. |
| Qubit dsDNA HS assay kit | Invitrogen | Q32851/Q32854 | step 3.6. |
| Qubit assay tubes | life technologies | Q32856 | step 3.6. |
| PBS pH 7.2 (1X) | Gibco | 20012-027 20012-050 | |
| Disposable scalpel No10 | Feather | 2975#10 | |
| Plastic K2 EDTA 2 ml tubes | BD vacutainer | 367841 | |
| syringe U-100 with 26G x ½” needle | BD vacutainer | 329652 | IP challenge, step 2.2.1. |
| 3 mL Luer-Lok tip syringe | BD vacutainer | 309657 | Cardiac puncture, step 2.4.2. |
| 25G X 5/8” needle | BD vacutainer | 305901 | Cardiac puncture, step 2.4.2. |
| 25 mm fritted glass base with stopper | EMD Millipore | XX1002502 | Filtration unit system, step 1.1.7. |
| 25 mm aluminum spring clamp | EMD Millipore | XX1002503 | Filtration unit system, step 1.1.7. |
| 15 ml borosilcate glass funnel | EMD Millipore | XX1002514 | Filtration unit system, step 1.1.7. |
| 125 ml side-arm Erlenmeyer flask | EMD Millipore | XX1002505 | Filtration unit system, step 1.1.7. |
| Acetate-cellulose filter VVPP (pore size 0.1 mm; diameter 25 mm) | EMD Millipore | VVLP02500 |
References
- Murray, G. L., et al. Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect Immun. 77 (2), 810-816 (2009).
- Adler, B., Lo, M., Seemann, T., Murray, G. L. Pathogenesis of leptospirosis: the influence of genomics. Vet Microbiol. 153 (1-2), 73-81 (2011).
- Marcsisin, R. A., et al. Use of a high-throughput screen to identify Leptospira mutants unable to colonize the carrier host or cause disease in the acute model of infection. J Med Microbiol. 62, 1601-1608 (2013).
- van Opijnen, T., Bodi, K. L., Camilli, A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 6 (10), 767-772 (2009).
- van Opijnen, T., Lazinski, D. W., Camilli, A. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol. 36, 1E.3.1-1E.3.24 (2015).
- Troy, E. B., et al. Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infect Immun. 81 (7), 2347-2357 (2013).
- Wunder, E. A., et al. Real-time PCR reveals rapid dissemination of Leptospira interrogans after intraperitoneal and conjunctival inoculation of hamsters. Infect Immun. 84 (7), 2105-2115 (2016).
- Lourdault, K., Aviat, F., Picardeau, M. Use of quantitative real-time PCR for studying the dissemination of Leptospira interrogans in the guinea pig infection model of leptospirosis. J Med Microbiol. 58, 648-655 (2009).
- Coutinho, M. L., et al. Kinetics of Leptospira interrogans infection in hamsters after intradermal and subcutaneous challenge. PLoS Negl Trop Dis. 8 (11), e3307 (2014).
- Athanazio, D. A., et al. Rattus norvegicus as a model for persistent renal colonization by pathogenic Leptospira interrogans. Acta Trop. 105 (2), 176-180 (2008).
- Ratet, G., et al. Live imaging of bioluminescent Leptospira interrogans in mice reveals renal colonization as a stealth escape from the blood defenses and antibiotics. PLoS Negl Trop Dis. 8 (12), e3359 (2014).
- Gutierrez, M. G., Yoder-Himes, D. R., Warawa, J. M. Comprehensive identification of virulence factors required for respiratory melioidosis using Tn-seq mutagenesis. Front Cell Infect Microbiol. 5 (78), (2015).
- Gawronski, J. D., Wong, S. M., Giannoukos, G., Ward, D. V., Akerley, B. J. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A. 106 (38), 16422-16427 (2009).
- Gallagher, L. A., Shendure, J., Manoil, C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2 (1), (2011).
- Slamti, L., Picardeau, M. Construction of a library of random mutants in the spirochete Leptospira biflexa using a mariner transposon. Methods Mol Biol. 859, (2012).
- Picardeau, M. Conjugative transfer between Escherichia coli and Leptospira spp. as a new genetic tool. Appl Environ Microbiol. 74 (1), 319-322 (2008).
- Lourdault, K., Matsunaga, J., Haake, D. A. High-throughput parallel sequencing to measure fitness of Leptospira interrogans transposon insertion mutants during acute infection. PLoS Negl Trop Dis. 10 (11), e0005117 (2016).
- Ellinghausen, H. C., McCullough, W. G. Nutrition of Leptospirapomona and growth of 13 other serotypes: Fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am J Vet Res. 26, 45-51 (1965).
- Johnson, R. C., Harris, V. G. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol. 94 (1), 27-31 (1967).
- Demarre, G., et al. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPa) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol. 156 (2), 245-255 (2005).
- Vallenet, D., et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford). , bap021 (2009).
- Goodman, A. L., Wu, M., Gordon, J. I. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat. Protocols. 6 (12), 1969-1980 (2011).
- Miller, J. . Spirochetes in body fluids and tissues manual of investigative methods. , 22-23 (1971).
- Ristow, P., et al. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3 (7), (2007).
- Croda, J., et al. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect Immun. 76 (12), 5826-5833 (2008).
- Haake, D. A. Hamster model of leptospirosis. Curr Protoc Microbiol. 12, 2 (2006).
- Giardine, B., et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15 (10), 1451-1455 (2005).
- Goecks, J., Nekrutenko, A., Taylor, J., Galaxy, T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11 (8), R86 (2010).
- Blankenberg, D., et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. (Chapter 19), 11-21 (2010).
- Afgan, E., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 8 (44), (2016).
- McCoy, K. M., Antonio, M. L., van Opijnen, T. MAGenTA; a Galaxy implemented tool for complete Tn-Seq analysis and data visualization. Bioinformatics. , 1367 (2017).
- Langmead, B., Trapnell, C., Pop, M., Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10 (3), R25 (2009).
- Kamp, H. D., Patimalla-Dipali, B., Lazinski, D. W., Wallace-Gadsden, F., Camilli, A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 9 (12), e1003800 (2013).
- Poggi, D., Oliveira de Giuseppe, P., Picardeau, M. Antibiotic resistance markers for genetic manipulations of Leptospira spp. Appl Environ Microbiol. 76 (14), 4882-4885 (2010).
- van Opijnen, T., Camilli, A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 11 (7), 435-442 (2013).
- Pappas, C. J., Picardeau, M. Control of gene expression in Leptospira spp. by Transcription activator-like effectors demonstrates a potential role for LigA and LigB in Leptospira interrogans virulence. Appl Environ Microbiol. 81 (22), 7888-7892 (2015).
- Koskiniemi, S., Sun, S., Berg, O. G., Andersson, D. I. Selection-driven gene loss in bacteria. PLoS Genet. 8 (6), e1002787 (2012).
- Troy, E. B., et al. Global Tn-seq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol Microbiol. 101 (6), 1003-1023 (2016).
- Ramsey, M. E., et al. A high-throughput genetic screen identifies previously uncharacterized Borrelia burgdorferi genes important for resistance against reactive oxygen and nitrogen species. PLoS Negl Trop Dis. 13 (2), e1006225 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved