Method Article
Optimized Interferon-gamma ELISpot Assay to Measure T Cell Responses in the Guinea Pig Model after Vaccination
In This Article
Summary
The development of the interferon-gamma ELISpot assay for guinea pig PBMCs allows the characterization of T-cell responses in this highly relevant model for studying infectious diseases. We have applied the assay to measure T cell responses associated with intradermal delivery of DNA vaccines.
Abstract
The guinea pig has played a pivotal role as a relevant small animal model in the development of vaccines for infectious diseases such as tuberculosis, influenza, diphtheria, and viral hemorrhagic fevers. We have demonstrated that plasmid-DNA (pDNA) vaccine delivery into the skin elicits robust humoral responses in the guinea pig. However, the use of this animal to model immune responses was somewhat limited in the past due to the lack of available reagents and protocols to study T cell responses. T cells play a pivotal role in both immunoprophylactic and immunotherapeutic mechanisms. Understanding T cell responses is crucial for the development of infectious disease and oncology vaccines and accommodating delivery devices. Here we describe an interferon-gamma (IFN-γ) enzyme-linked immunospot (ELISpot) assay for guinea pig peripheral blood mononuclear cells (PBMCs). The assay enables researchers to characterize vaccine-specific T-cell responses in this important rodent model. The ability to assay cells isolated from the peripheral blood provides the opportunity to track immunogenicity in individual animals.
Introduction
The protocol described here permits the detection of interferon-gamma (IFN-γ) secreting cells after antigen recall in a peripheral blood mononuclear cell (PBMC) population harvested from Hartley guinea pigs. We have applied the assay to characterize the kinetics and magnitudes of antigen-specific T cell responses to an Influenza vaccination-regimen in the guinea pig. We believe this protocol will significantly propel the pre-clinical development of vaccination programs in this highly relevant animal model.
T cells elicited by vaccines play an essential role in the protection against infectious agents and immunotherapeutic pathways associated with other diseases. The importance of T cells has been highlighted in multiple vaccine studies. Immunization of ferrets and mice with a plasmid DNA (pDNA) encoding for H5 hemagglutinin and N1 neuraminidase provided protection from morbidity and mortality in an influenza virus challenge in the absence of neutralizing antibodies, indicating the importance of T cell immunity1. In addition to strong neutralizing humoral response, T-cells play a crucial role for not only viral clearance2 but also protection from infection with respiratory syncytial virus (RSV) in mice3. In humans, pre-existing CD8+ T cells were associated with decreased disease severity during the H1N1 pandemic in 20094. CD4+ T cell counts together with certain cytokine plasma concentration are correlated with disease severity in RSV-infected children5.
The guinea pig has gained prominence as a laboratory model for research and development in various areas of medicine such as skin sensitization, nutritional research, studies of the auditory system and, most relevant for this work, infectious diseases. It was crucial for the discovery of vaccines against tuberculosis and diphtheria. More recently the guinea pig is used as a model for Influenza6 and Ebola7. Furthermore, possessing physiological similarities to human skin8, the guinea pig offers an accessible small animal model for dermal drug delivery methods. In contrast to its importance as a laboratory model, the availability of guinea pig specific assays and probes to characterize immune responses remains limited9. Basic cellular assays such as the IFN-γ enzyme-linked immunospot (ELISpot-assay) that is routinely used in pre-clinical and clinical research to enumerate T cell responses have not been available.
The first solid-phase enzyme-linked immunospot assays were used to determine the number of specific antibody-secreting cells in a diverse B cell-population10,11. The format has advanced to detect cells secreting cytokines including IL-1, IL-2, IL-4, IL-5, IL-6, IL-10, GM-CSF, TNF-alpha, TNF-beta, granzyme B, and IFN-γ. The ELISpot assay possesses high sensitivity; potentially each cytokine-producing cell can be detected. The limit of detection for ELISpot assays has been reported to be lower than 10 spots per 100,000 PBMCs12. Recently a guinea-pig IFN-γ specific antibody pair was made available13, and this has opened up the opportunity to address this deficiency in the field.
IFN-γ is considered an important effector molecule in the adaptive immune response; it can be produced and secreted by both CD4 and CD8 T cells upon activation. IFN-γ has a wide range of biological functions in the context of an immune response to a virus infection, such as the activation of macrophages and the up regulation of major histocompatibility complex (MHC) I and MHCII expression. It also promotes B-cell differentiation, inhibits T-helper-2 cell growth, and activates natural killer cells. IFN-γ blocks viral replication in infected somatic cells and upregulates expression of MHC molecules. Thus, the production of IFN-γ is a very important indication of the quality of the T cell response to a vaccine or pathogen.
An important aspect of the here presented IFN-γ ELISpot is the use of PBMCs rather than splenocytes13. PBMCs can be obtained by processing a non-terminal collected blood sample, whereas the collection of splenocytes requires animal euthanization before harvesting of the spleen. The use of PBMCs enables IFN-γ T-cell responses to be monitored over a period of time and evaluation of the effects on the T-cell responses such as prime-boost regimens14.
For researchers who are currently using the guinea pig as an animal model, the IFN-γ ELISpot method will broaden the range of scientific data they can gain. Its availability may now reduce the necessity to conduct studies with less relevant animal models for the target disease, which were previously chosen due to reagent availability for examination of cellular responses. The use of PBMCs rather than spleen or lymph nodes allows for non-terminal experiments and continuous monitoring of individual animals.
Below we provide a detailed description of the steps involved in the detection of an IFN-γ cellular response in guinea pigs to pDNA vaccine. We outline our specific delivery procedure and describe the general ELISpot assay encompassing blood sampling, blood processing, PBMC harvest, assay procedure, and data analysis. A schematic of the ELISpot assay is depicted in Figure 1.
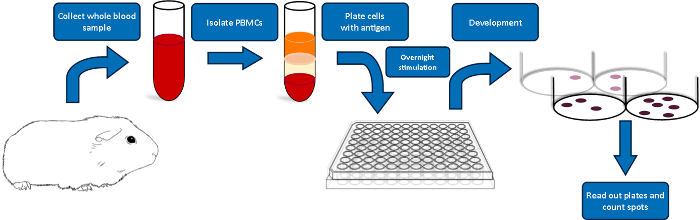
Figure 1: Schematic overview of the guinea pig ELISpot protocol. Please click here to view a larger version of this figure.
Protocol
All methods described here have been approved by the Animal Care and Use Committee (ACUC) of Acculab.
NOTE: The protocol requires the use of potential hazardous materials. Please refer to manufacturer's MSDS and wear proper personal equipment (PPE) throughout the procedure.
1. Preparation of the guinea-pig immunization site, intradermal Mantoux-injection and CELLECTRA-3P-EP-procedure
- Preparation of the treatment site on the guinea pig.
- Place guinea pig in an induction chamber and anaesthetize at 5% Isoflurane-vapor.
- Remove animal from the chamber and place nose cone in position, providing 2% Isoflurane vapor to maintain anesthesia throughout the procedure.
- Shave area of approximately 4 cm2 on abdominal flank.
- Disinfect shaved area with Ethanol-swab.
- Injection of pDNA-formulation and electroporation procedure.
- Use an Insulin-syringe to inject 100 µl formulation into the dermis by the Mantoux technique. Needle is inserted bevel up at a 10 degree angle with the animal’s body.
- Remove the needle and immediately insert the CELLECTRA®-3P electrode array across the injection-wheal and apply electrical field.
- Remove animal from anesthesia and monitor its recovery.
2. Non-terminal bleed
- Blood collection from jugular vein of anaesthetized guinea pig.
- Place guinea pig in an induction chamber and anaesthetize animal as described in 1.1.1
- Using a 3 ml syringe with a 27G 1/2inch needle, punctuate the jugular vein of the anaesthetized animal and collect 3 ml of blood.
- Immediately transfer blood into K2EDTA blood collection tube, invert tube several times to mix blood with anti-coagulant and place on ice.
NOTE: Avoidance of blood clotting is crucial for successful downstream processing of the blood sample. - Monitor animal’s recovery.
3. Blood sample processing
- Separation of PBMCs from whole blood by density-gradient centrifugation
- Dilute blood 1:1 with Hank’s Balanced Salt Solution (HBSS).
NOTE: To avoid contamination all work from here on should be performed in a Biosafety cabinet applying aseptic technique. - Layer diluted blood gradually over 4.5 ml Ficoll-Paque Plus in a 15 ml-conical tube. Avoid disturbance of the interface. NOTE: To ensure proper separation, Ficoll-Paque must be at Room temperature.
- Centrifuge tubes at 800 rcf for 30 min with brake off.
NOTE: As an alternative, SepMate™ tubes (STEMCELL Technologies) can be used for PBMC-separation. HBSS-diluted blood is added to 15 ml SepMate tube filled with 3.5 ml Ficoll-Paque Plus and then centrifuged at 1200 rcf for 10 min (brake on). This time-saving gradient centrifugation technique results in equal viability, cell-yield and spot-formation. - Harvest Buffy-Coat layer and dilute in R10 medium (10% (v/v) heat-deactivated FBS and 1% (v/v) Pen/Strep in RPMI1640 medium) to a total volume of 15 ml.
- Pellet cells by centrifuging at 450 rcf for 5 min.
- Wash cells twice with R10 medium.
- Re-suspend cells in 1 ml R10 and pass cell-suspension through 70 µm cell-strainer in to a new tube.
- Count cells and determine number of live cells by Trypan-blue staining.
- Dilute cells with R10 medium to a concentration of 1x10^6 live cells per ml.
- Dilute blood 1:1 with Hank’s Balanced Salt Solution (HBSS).
4. Preparation of ELISpot-plates and cell-stimulation
- Coat and block wells of a 96-well ELISpot PVDF-membrane plate before seeding the PBMCs.
- Plates should be pre-treated for 60 seconds. Ethanol pre-treatment will result in less unspecific spot-formation and spots with improved definition.
NOTE: Take extra care to ensure the complete membrane comes into contact with 15 µl 35% Ethanol. - After 60 seconds add 150 µl 1x PBS per well, and then empty out wells by inverting the plates.
NOTE: The Ethanol pre-treatment is very time-sensitive. Do not incubate the well-membrane longer than 60 sec. Membranes must not dry out during the following steps of the procedure. - Wash plates three times with 250 µl 1x PBS per well.
- Coat with 100 µl/well of 5 µg/ml capture anti-guinea pig IFN-γ antibody V-E4 in PBS.
- Incubate plates for at least 12 hrs at 4° C.
- Wash plates by adding 250 µl/well PBS three times.
- Add 200 µl/well blocking buffer (10% (w/v) Sucrose and 2% (w/v) BSA in PBS) and incubate for 2 hours at room temperature.
- Wash plates with 250 µl/well PBS three times.
- Plates should be pre-treated for 60 seconds. Ethanol pre-treatment will result in less unspecific spot-formation and spots with improved definition.
- Plate PBMCs with antigen-specific peptides, positive and negative controls.
- Dilute protein-peptide pool in R10 to desired final peptide concentration.
- Assay the sample PBMCs in triplicates.
NOTE: Add medium with stimulants to the ELISpot plates first and add the PBMC suspension second. - Add 50 µl of peptide-R10 medium to indicated wells.
- For negative control add 50 µl empty peptide formulation in R10 into indicated wells.
- For positive control add 5 µg/ml Concanavalin A (ConA) in R10 medium into indicated wells.
- Add 100 µl of PBMC cell-suspension to all wells
- Incubate plates in humidified 5% CO2 atmosphere at 37° Celsius for 18 hrs.
NOTE: Place ELISpot plates on an even surface/rack in the incubator and avoid any disturbance of the plates during the incubation time.
5. Detection of Interferon-gamma positive spots
- Incubation with detection Antibody and plate development.
NOTE: For all steps in this section no aseptic technique is required.- Carefully remove plates from incubator, and safely empty wells. Wash Plates by adding 250 µl/well PBS for three times.
- Add 100 µl/well of 2 µg/ml biotinylated detection anti-guinea pig IFN-γ antibody N-G3 diluted in blocking buffer.
NOTE: Filter (22 µm) antibody solution to reduce background. - Incubate with detection antibody for 2 hrs at room temperature.
- Discard supernatant and wash plates by adding 250 µl/well PBS three times.
- Add 100 µl/well of alkaline phosphatase (ALP)-conjugated streptavidin diluted in blocking buffer.
NOTE: Filter (22 µm) solution to reduce background. - Incubate with ALP-Streptavidin conjugate for 1 hr at room temperature.
- Discard ALP-Streptavidin solution and wash plates by adding 250 µl/well PBS two times, and remove excess PBS by inverting the plate and blotting it against a clean paper towel.
- Wash plates by adding 250 µl/well UltraPure DI water one time, and remove excess water by inverting the plate and blotting it against a clean paper towel.
- Add 100 µl of BCIP/NBT (CAUTION) substrate solution into each well. Caution: BCIP/NBT is highly flammable and toxic if swallowed, in contact with skin, or inhaled. Before use refer to the MSDS.
- Incubate for 20 min at room temperature protected from light.
- Rinse the plate 4 times with deionized water. Invert plate and tap to remove excess water.
- Remove plastic drainage from bottom of ELISpot plates and allow the plates to dry.
- Quantify spots manually or by using an automated ELISpot reader, for example the CTL-Immunospot S6 plate reader.
Results
The results presented here serve as a reference for expected outcomes following the use of this protocol, emphasizing the importance of crucial steps and confirming the benefits of described optimizations.
After density-gradient centrifugation, as described in step 3.1 of the protocol, the red viscous liquid at the bottom of the tube will contain most of the red blood cells. As shown in Figure 2A, above the red blood cells is a layer of the density-medium. Between a layer of clear or yellow plasma on top and the density gradient medium is the white or light brown buffy coat layer, which contains most of the white blood cells and platelets. Incomplete separation would manifest as the presence of red blood cells on top of the density-gradient medium. The red coloration of the plasma in Figure 2A is most probably due to hemolysis, as this blood sample was stored in the collection tube 1.5 h before being processed. Figure 2B shows the gradient before (left) and after (right) centrifugation in PBMC-isolation device. Blood sample in Figure 2B was processed with minimal delay after blood collection, and the plasma coloration is yellow.
The effect of pre-wetting the membranes with ethanol (see step 4.1 of the protocol) on spot-development is shown in Figure 3. Spots in pre-treated wells display improved definition, and there is a reduction in the number of spots in medium/DMSO negative control wells (Figure 3A). Typical viability of processed guinea pig PBMCs ranges around 90% and is similar for both regular 15 mL tubes and PBMC-isolation devices (Figure 3B). Yield of viable cells from a 3 mL blood sample typically ranges between 3 x 106 and 5 x 106.
Animals were immunized with plasmid DNA encoding the nucleoprotein (NP) of H1N1 Influenza strain A/PuertoRico8. Figure 4 shows enumeration of the T-cell IFN-γ responses presented as spot-forming units (SFU) generated across the duration of a vaccination regimen. ELISpot assay with PBMCs harvested from non-immunized animals results in negligible spot counts (no tx). Fourteen days after the first immunization (prime), an average of 970 IFN-γ spots per million PBMCs were counted after immunogenic stimulation. Seven days after the second immunization (boost), T-cell responses against all three pools were expanded, reaching an average of 5020 IFN-γ SFUs/106 PBMCs. Forty-six days after the second immunization (memory), an average total of 6310 IFN-γ SFUs/106 PBMCs were counted in peripheral blood.
FIGURE LEGENDS:
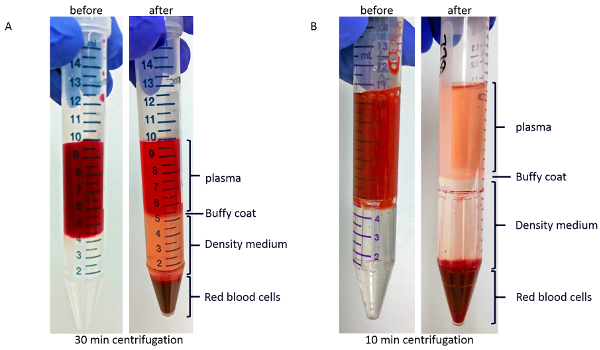
Figure 2: Representative images of layers after density-gradient centrifugation. (A) Blood sample before (left) and after (right) centrifugation in regular tubes. (B) Blood sample before (left) and after (right) centrifugation in PBMC-isolation devices. Please click here to view a larger version of this figure.
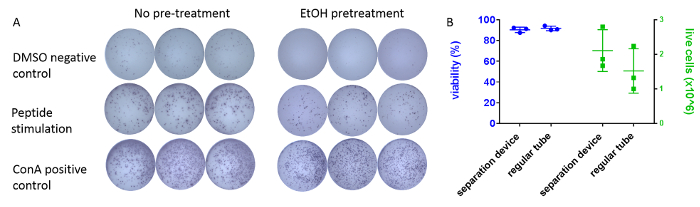
Figure 3: Optimization of the protocol. (A) Typical appearance of IFN-γ positive spots in an ELISpot assay plate well developed according to the protocol. Example triplicate wells are shown after pre-treatment with ethanol (right) or without pre-treatment (left). Cells were incubated overnight either with DMSO (top) as no-stimulus control, antigen-matched peptides (middle), or the mitogen ConA (bottom). PBMCs originated from the same individual animal. Wells were imaged using an automated plate-scanner. (B) Comparison of different density-gradient tubes on processed cells. 3 mL of blood samples from 3 individual guinea pigs were collected. 1.5 mL of each sample were processed using either regular tubes (density gradient medium) or PBMC-isolation devices. Viability (left, % ± SEM) and number of live cells (right, x 106 ± SEM) were determined using an automatic cell counting system and trypan-blue exclusion assay. Please click here to view a larger version of this figure.
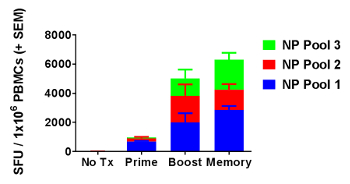
Figure 4: Detection of robust cellular immune responses over the course of a vaccination regimen. On days 1 and 15, 30 µg of pDNA encoding influenza A nucleoprotein (NP) was delivered intra-dermally to abdominal flank of Hartley guinea pigs immediately followed by skin-electroporation. IFN-γ ELISpot response was measured 14 days after the first immunization (prime), and 7 days (boost) and 46 days (memory) after the second immunization. Mean SFUs + SEM were plotted for 5 treated guinea pigs and 2 untreated guinea pigs (no tx). Please click here to view a larger version of this figure.
Discussion
The guinea pig is a valuable animal model for the pre-clinical development of vaccines and intradermal delivery strategies. The above protocol describes the methodology to measure antigen-specific T cell responses in this highly relevant model. The assay provides a clear enumeration of peripheral T-cells producing Interferon-gamma upon stimulation with antigen-specific peptides. The kinetics of the immune response can be monitored by non-terminal blood sampling.
We optimized the protocol and identified critical aspects to obtain optimal results using this assay. For example, the formation of blood clots in the collection tube will result in suboptimal PBMC recovery and viability. Guinea pig blood clots very rapidly18. It's crucial to perform the blood draw quickly. Au-Birck et al. provide a comprehensive guide for bleeding techniques in the guinea-pig model16. Since the blood collection requires general anesthesia, it is recommended to review applicable standard operating procedures of this aspect of the procedure19. Insufficiently anaesthetized guinea pigs will react by moving their legs or vocalization after a brief pinch of the tissue between their toes with your fingernails or by flinching their ears and moving their whiskers forward after pinching their ear. The immediate transfer of the blood into a tube with anticoagulant and mix thoroughly by rolling or inverting the tube several times is an essential step in this protocol. For the here described protocol EDTA-tubes were used, and no other anticoagulants were tested. Please note that EDTA is hypertonic, tubes should ideally be filled more than half full, and therefore the appropriate tube size for the sample volume must be used.
When performed correctly, density gradient centrifugation consistently results in clean PBMC preparation. The density-gradient medium must be at room temperature when layering the gradient before centrifugation, and brakes should not be used for stopping the centrifuge when using regular tubes. A significant improvement in terms of practicality of the PBMC-processing was the combination of density gradient centrifugation with PBMC-isolation devices. This allows for a reduction in centrifugation time. Density gradient centrifugation in PBMC-isolation devices and regular tubes results in similar viability, live cell counts of processed PBMCs, and spot formation. Independent of the type of tube used for the separation, the buffy coat harvest should follow immediately. Over an extended period of time contact with the density gradient medium is cytotoxic to the PBMCs.
Accurate counting of viable PBMCs is important to seed consistently equal numbers of cells into the assay-plate wells. Some method of live/dead discrimination should be applied. In our lab, we use the trypan-blue exclusion assay and an automated cell-counter.
Spot quality, defined as sharp-edged and high contrasted spots, will result in improved accuracy and consistent spot counting, especially when using an automated counting system. In line with the manufacturer's recommendations, we observed ethanol pre-treatments of the ELISpot plates to be a crucial step to reduce the number of background spots and improve the definition of the spots. The use of a plate reader to image the ELISpot assay-plates at the end of the protocol is highly recommended. Original images of each well can be easily and efficiently obtained and stored. Using an image analysis software digital images also allow for objective analysis and spot counting compared to manual counting by individual operators.
An absolute necessity for the development of the assay described here was the generation of a suitable anti-guinea pig Interferon-gamma antibody pair. Mouse monoclonal antibodies V-E4 and N-G3 were developed by hybridoma-technique with B-cell clones from mice which were immunized with recombinant guinea-pig interferon gamma20. V-E4, which is IgG1 isotype, and N-G3, which is IgG2a, are reported to bind both to the recombinant and native antigen. Assigning V-E4 as the capture antibody and biotinylated N-G3 as the detection antibody resulted in high sensitivity for both recombinant and native protein while retaining low background signal in a sandwich-ELISA format. Both antibodies are available to the scientific community through a manufacturing agreement led by the laboratory of Dr. Hubert Schaefer, who is co-author of this publication. Requests will be received by Dr. Schaefer's lab and then manufactured by a commercial partner.
Interferon-gamma is reported to be unstable in regular buffers or media21. Schaefer et al.20 reported only a moderate decrease of recovery rate when using degraded IFN-y, which had lost biological functionality, in an ELISA format using antibodies V-E4 and N-G3. However, increasing the incubation time of peptide stimulation of PBMCs over 18 h should be carefully tested.
The advantages of using PBMCs has been discussed. However, the here described assay only reflects antigen-specific responses of circulating T-cells in the periphery. Tissue-infiltrating T-cells or cells isolated from lymphatic organs, such as spleen and lymph nodes, may exhibit different properties22,23,24. No significant differences in cellular responses were observed upon comparison of interferon-gamma spots from PBMCs and splenocytes from guinea pigs that were immunized with the same pNP influenza vaccine14.
In this protocol, we described an intradermal vaccination procedure. A principal rationale for ID immunization is targeting the high density of dendritic cells present in the skin25. We and others have shown that these cells can be specifically targeted by adapting delivery-method26, formulation27, or drug-design28. Activated professional antigen-presenting cells may migrate to a draining lymph node and activate the adaptive immune system29,30,31,32. Dendritic cells are the essential antigen presentation cell-type for priming productive cellular immune responses.
For this study animals were immunized with plasmid DNA encoding the nucleoprotein of influenza H1N1 strain A/PuertoRico/8. Skin delivery of this pDNA vaccine (pNP) in combination with electroporation had been shown to elicit antigen-specific humoral responses in guinea pigs33, ferrets, and non-human primates (NHPs)1,34. Additionally, this vaccine elicited robust T-cell responses in mice after delivery into the epidermis35, which could be attributed to CD4 and CD8 T-cells by stimulating with peptides representing specific epitopes for these cell-populations. The pNP vaccine was also immunogenic after mucosal delivery as demonstrated by generation of humoral responses in rabbits and guinea pigs, as well as cellular and humoral responses in mice36. Most recently, our group was able to demonstrate the generation of IFN-γT-cell responses in the rabbit after intra-muscular delivery (unpublished data).
Currently, the observed Interferon-gamma spots in the guinea pig ELISpot cannot be allocated to a CD4+ or CD8+ T-cell subset. Especially for the design and development of immune therapies targeting the skin, it would be very useful to determine whether observed interferon-gamma spot frequencies are caused by expansion of one particular subset or a balanced response of both in order to evaluate the effectiveness of the vaccination to trigger cross-presentation. This limitation might be overcome by negative cell-sorting for CD4 or CD8 prior to seeding the cells on the plate or by identification of MHC class II (for CD4 responses) or MHC class I (for CD8 responses) restricted epitopes.
The here described Interferon-gamma ELISpot assay using guinea pig PBMCs addresses the need to assess the course of cellular responses in the guinea pig laboratory model. This will refine the development of vaccines and skin delivery protocols. We believe that it allows for the employment of this relevant animal model to study diseases with important T-cell components such as TB37, Ebola38, HSV39, and others. It will reduce the use of less relevant animal models.
Disclosures
The authors Katherine Schultheis, Holly M. Pugh, Bryan S. Yung, Janet Oh, Holly M. Pugh, Jacklyn Nguyen, Laurent Humeau, Kate E. Broderick, and Trevor R.F. Smithare employees of Inovio Pharmaceuticals and as such receive salary and benefits, including ownership of stock and stock option, from the company.
Acknowledgements
We would like to thank the members of the Inovio Pharmaceuticals R&D department for technical assistance and staff of Acculab for providing excellence husbandry service. In particular we would like to thank Alysha Vu and Joe Agnes for proofreading the manuscript. This work was not funded by any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Materials
| Name | Company | Catalog Number | Comments |
| Cellectra-3P | Inovio Pharmaceuticals | ID-Electroporatio device/ non-commercial, R&D-only | |
| K2EDTA blood collection tube | BD | 367844 | |
| Hank’s Balanced Salt Solution | Gibco | 14175-079 | |
| Ficoll-Paque Plus | GE Healthacre | 17144003 | density gradient medium |
| SepMate tube | STEMCELL Technologies | 85415 | PBMC-isolation device |
| RPMI | Thermo Fisher | 11875-135 | |
| heat-deactivated FBS | VWR | 97068-085 | |
| Pen/Strep | Gibco | 15140-122 | |
| β-Mercaptoethanol | Gibco | 21985-023 | |
| 96-well ELISpot PVDF-membrane | Millipore | MSIPS4W10 | ELISpot assay plate |
| Ethanol | Sigma | 459844-1L | |
| UPDI | Invitrogen | 10977-015 | Ultra-Pure DI water |
| Sucrose | Sigma | S7903 | |
| Bovine Serum Albumin | Sigma | A7906 | |
| 10x PBS | HyClone | SH30378.03 | |
| Concanavalin A (ConA) | Sigma | C5275-5MG | |
| alkaline phosphatase (ALP)-conjugated streptavidin | R&D Systems Inc. | SEL002 | |
| BCIP/NBT | R&D Systems Inc. | SEL002 | |
| capture anti-guinea pig IFN-γ antibody V-E4 | GenScript | Order #:U5472CE080-3 | LOT # A217071153 |
| biotinylated detection anti-guinea pig IFN-γ antibody N-G3 | GenScript | Order #:U5472CE080-1 | LOT # A21700598 |
| CTL S6 Micro Analyzer | ImmunoSpot | #S6MIC12 | automated ELISpot plate reader |
| Vi-CELL XR | Beckman-Coulter | 731050 | automated cell counter |
| ImmunoSpot 5.1 | ImmunoSpot | assay analysis software | |
| ESCO Class II Type A2 | ESCO | Biosafety cabinet | |
| Falcon cell strainer | Corning, Inc. | VWR cat# 21008-952 | |
| Exel Comfort Point Insulin syringe | MedLab Supply | 26028 | |
| Rotanta 460R | Hettich | centrifuge | |
| Vi-CELL XR Quad Pak Reagent Kit | Beckman Coulter | 383722 | |
| Incu Safe | Panasonic-Healthcare | incubator | |
| 22 µm Filter | ThermoScientific | VWR act# 597-4520 | 22 µm Filter |
| KimWipes | Kimberly-Clark Professional | VWR cat# 21903-005 | paper towels |
References
- Laddy, D. J., et al. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PloS One. 3 (6), 2517 (2008).
- Lee, J. Y., Chang, J. Recombinant baculovirus-based vaccine expressing M2 protein induces protective CD8+ T-cell immunity against respiratory syncytial virus infection. Journal of Microbiology. 55 (11), 900-908 (2017).
- Kinnear, E., et al. Airway T cells protect against RSV infection in the absence of antibody. Mucosal Immunology. , (2017).
- Sridhar, S., et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nature Medicine. 19 (10), 1305-1312 (2013).
- Brand, H. K., et al. CD4+ T-cell counts and interleukin-8 and CCL-5 plasma concentrations discriminate disease severity in children with RSV infection. Pediatric Research. 73 (2), 187-193 (2013).
- Bouvier, N. M. Animal models for influenza virus transmission studies: a historical perspective. Current Opinion in Virology. 13, 101-108 (2015).
- St Claire, M. C., Ragland, D. R., Bollinger, L., Jahrling, P. B. Animal Models of Ebolavirus Infection. Comparative Medicine. 67 (3), 253-262 (2017).
- Todo, H. Transdermal Permeation of Drugs in Various Animal Species. Pharmaceutics. 9 (3), 33 (2017).
- Schafer, H., Burger, R. Tools for cellular immunology and vaccine research the in the guinea pig: monoclonal antibodies to cell surface antigens and cell lines. Vaccine. 30 (40), 5804-5811 (2012).
- Czerkinsky, C. C., Nilsson, L. A., Nygren, H., Ouchterlony, O., Tarkowski, A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. Journal of Immunological Methods. 65 (1-2), 109-121 (1983).
- Sedgwick, J. D., Holt, P. G. A solid-phase immunoenzymatic technique for the enumeration of specific antibody-secreting cells. Journal of Immunological Methods. 57 (1-3), 301-309 (1983).
- Moodie, Z., et al. Response definition criteria for ELISPOT assays revisited. Cancer Immunology, Immunotherapy. 59 (10), 1489-1501 (2010).
- Gillis, P. A., et al. Development of a novel, guinea pig-specific IFN-gamma ELISPOT assay and characterization of guinea pig cytomegalovirus GP83-specific cellular immune responses following immunization with a modified vaccinia virus Ankara (MVA)-vectored GP83 vaccine. Vaccine. 32 (31), 3963-3970 (2014).
- Schultheis, K., et al. Characterization of guinea pig T cell responses elicited after EP-assisted delivery of DNA vaccines to the skin. Vaccine. 35 (1), 61-70 (2017).
- . Mantoux Tuberculin Skin Test DVD Facilitator Guide - CDC (Part One) Available from: https://www.cdc.gov/tb/education/Mantoux/images/mantoux.pdf (2013)
- Birck, M. M., Tveden-Nyborg, P., Lindblad, M. M., Lykkesfeldt, J. Non-Terminal Blood Sampling Techniques in Guinea Pigs. Journal of Visualized Experiments. (92), e51982 (2014).
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Current Protocols in Immunology. 111, 1-3 (2014).
- Lewis, J. H. Comparative hematology: studies on guinea-pigs (Cavia porcellus). Comparative Biochemistry and Physiology: Comparative Physiology. 102 (3), 507-512 (1992).
- Flecknell, P., Flecknell, P. . Laboratory Animal Anaesthesia (Fourth Edition). , 77-108 (2016).
- Schaefer, H., Kliem, G., Kropp, B., Burger, R. Monoclonal antibodies to guinea pig interferon-gamma: tools for cytokine detection and neutralization. Journal of Immunological Methods. 328 (1-2), 106-117 (2007).
- Lipiainen, T., et al. Formulation and stability of cytokine therapeutics. Journal of Pharmaceutical Sciences. 104 (2), 307-326 (2015).
- Wang, X. Z., et al. Virus-Specific CD8 T Cells in Peripheral Tissues Are More Resistant to Apoptosis Than Those in Lymphoid Organs. Immunity. 18 (5), 631-642 (2003).
- Veron, P., et al. Deep Sequencing of T Cell Receptor in Peripheral Blood and Muscle from Adeno-Associated Virus Vector-Injected Subjects Reveals Differences in T Cell Clonality Between the Two Compartments. Molecular Therapy. 23, 210 (2015).
- Sckisel, G. D., et al. Differential phenotypes of memory CD4 and CD8 T cells in the spleen and peripheral tissues following immunostimulatory therapy. Journal for ImmunoTherapy of Cancer. 5 (1), 33 (2017).
- Yanofsky, V. R., Mitsui, H., Felsen, D., Carucci, J. A. Understanding Dendritic Cells and Their Role in Cutaneous Carcinoma and Cancer Immunotherapy. Clinical and Developmental Immunology. 2013, 624123 (2013).
- Amante, D. H., et al. Direct Transfection of Dendritic Cells in the Epidermis After Plasmid Delivery Enhanced by Surface Electroporation. Human Gene Therapy Methods. 25 (6), 315-316 (2014).
- Mahe, B., et al. Nanoparticle-based targeting of vaccine compounds to skin antigen-presenting cells by hair follicles and their transport in mice. Journal of Investigative Dermatology. 129 (5), 1156-1164 (2009).
- Vandermeulen, G., et al. Skin-specific promoters for genetic immunisation by DNA electroporation. Vaccine. 27 (32), 4272-4277 (2009).
- Brave, A., Nystrom, S., Roos, A. K., Applequist, S. E. Plasmid DNA vaccination using skin electroporation promotes poly-functional CD4 T-cell responses. Immunology and Cell Biology. 89 (3), 492-496 (2011).
- Romani, N., et al. Targeting skin dendritic cells to improve intradermal vaccination. Current Topics in Microbiology and Immunology. 351, 113-138 (2012).
- Smith, T. R., et al. DNA vaccination strategy targets epidermal dendritic cells, initiating their migration and induction of a host immune response. Molecular Therapy Methods & Clinical Development. 1, 14054 (2014).
- Teunissen, M. B., Haniffa, M., Collin, M. P. Insight into the immunobiology of human skin and functional specialization of skin dendritic cell subsets to innovate intradermal vaccination design. Current Topics in Microbiology and Immunology. 351, 25-76 (2012).
- Lin, F., et al. A novel prototype device for electroporation-enhanced DNA vaccine delivery simultaneously to both skin and muscle. Vaccine. 29 (39), 6771-6780 (2011).
- Laddy, D. J., et al. Electroporation of synthetic DNA antigens offers protection in nonhuman primates challenged with highly pathogenic avian influenza virus. Journal of Virology. 83 (9), 4624-4630 (2009).
- Lin, F., et al. Optimization of Electroporation-Enhanced Intradermal Delivery of DNA Vaccine Using a Minimally Invasive Surface Device. Human Gene Therapy Methods. 23 (3), 157-168 (2012).
- Kichaev, G., et al. Electroporation mediated DNA vaccination directly to a mucosal surface results in improved immune responses. Human Vaccines & Immunotherapeutics. 9 (10), 2041-2048 (2013).
- Klunner, T., Bartels, T., Vordermeier, M., Burger, R., Schafer, H. Immune reactions of CD4- and CD8-positive T cell subpopulations in spleen and lymph nodes of guinea pigs after vaccination with Bacillus Calmette Guerin. Vaccine. 19 (15-16), 1968-1977 (2001).
- Shedlock, D. J., et al. Induction of Broad Cytotoxic T Cells by Protective DNA Vaccination Against Marburg and Ebola. Molecular Therapy. 21 (7), 1432-1444 (2013).
- Hensel, M. T., et al. Prophylactic Herpes Simplex Virus 2 (HSV-2) Vaccines Adjuvanted with Stable Emulsion and Toll-Like Receptor 9 Agonist Induce a Robust HSV-2-Specific Cell-Mediated Immune Response, Protect against Symptomatic Disease, and Reduce the Latent Viral Reservoir. Journal of Virology. 91 (9), 02257 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved