In Silico Modeling Method for Computational Aquatic Toxicology of Endocrine Disruptors: A Software-Based Approach Using QSAR Toolbox
In This Article
Summary
Quantitative structure-activity relationship (QSAR) modeling is a representative bioinformatics-assisted method in toxicological screening. This protocol demonstrates how to computationally assess the risks of endocrine disruptors (EDs) in aquatic environments. Utilizing the OECD QSAR Toolbox, the protocol implements an in silico assay for analyzing toxicity of EDs in fish.
Abstract
Computational analyses of toxicological processes enables high-throughput screening of chemical substances and prediction of their endpoints in biological systems. In particular, quantitative structure-activity relationship (QSAR) models have been increasingly applied to assess the environmental effects of a plethora of toxic materials. In recent years, some more highlighted types of toxicants are endocrine disruptors (EDs, which are chemicals that can interfere with any hormone-related metabolism). Because EDs may significantly affect animal development and reproduction, rapidly predicting the adverse effects of EDs using in silico techniques is required. This study presents an in silico method to generate prediction data on the effects of representative EDs in aquatic vertebrates, particularly fish species. The protocol describes an example utilizing the automated workflow of the QSAR Toolbox software developed by the Organization for Economic Co-operation and Development (OECD) to enable acute ecotoxicity predictions of EDs. As a result, the following are determined: (1) calculation of the numerical correlations between the concentration for 50% of lethality (LC50) and octanol-water partition coefficient (Kow), (2) output performances in which the LC50 values determined in experiments are compared to those generated by computations, and (3) the dependence of estrogen receptor binding affinity on the relationship between Kow and LC50.
Introduction
New developments in informatics and computational technology have empowered the biological sciences with quantitative methodologies that offer high precision and reliability1. In particular, algorithms used in molecular taxonomy and property classification have resulted in quantitative structure-activity relationship (QSAR) models2. These models automatically correlate the chemical structures and biological activities of a given chemical database and implement rapid in silico screening of a wide range of chemical substrates according to their medicinal or toxicological actions3. QSAR tools can produce predictive toxicity profiles as a function of feature vectors of molecular descriptors (i.e., physicochemical parameters) of chemicals of interest to numerically create categorical endpoints4. Usually, each quantitative endpoint is displayed as a 2D scatterplot vs. changes in descriptor values. A QSAR model is then generated using (multiple) linear regression analyses. Once a dataset has been fully exploited to construct a QSAR model (called the training set), then the model is statistically validated by predicting the endpoints of a group of chemicals not included in the training set (called the test set). The model can then be used to predict the biological activities of untested compounds3.
Among many harmful chemicals, endocrine disruptors (EDs) have been highlighted as a group of toxicants that may interfere in numerous hormone-related metabolisms in mammals, amphibians, and fish5,6. EDs are known to induce a variety of adverse effects, such as cancers and malformations, by blocking or altering normal hormonal pathways or activating abnormal hormone synthesis/degradation signals. As a consequence, these hormone-mimicking chemicals can perturb endocrine systems such that biological development and reproduction of wildlife animal populations are hampered. In particular, the ecotoxicological effects of EDs have been extensively investigated in aquatic vertebrates, which have nearly identical hormone receptor structures to those of mammals, including humans. Because all hormonal actions occur at low doses in vivo, predicting the potential toxicities of ED candidates using rapid in silico screening is critical to public and environmental health.
QSAR models based on the toxicology of EDs have been conducted utilizing both 2D and 3D descriptors (known as 2D and 3D QSAR, respectively), which reveal the ED ligand binding affinities of estrogen, androgen, and progesterone receptors7. Despite the high-precision advantages of 3D QSAR, in which conformational and electrostatic interactions are considered, 2D QSAR retains its own robustness in direct mathematical algorithms, rapid calculations, and extremely low computational loads. In addition, 2D-QSAR models are flexible for use in a wide range of applications while achieving relatively accurate prediction performance.
The OECD QSAR Toolbox is currently one of the most utilized computer software tools, providing freely available and pre-built QSAR models8,9. Its profiler uses 2D descriptor databases. Since the release of the first version in 2008, the software has been applied in the fields of chemical and biological industries, public health, and environmental safety for full or partial analysis of the potential risks of natural and synthetic compounds, with special interests in carcinogenesis10,11,12, mutagenicity13,14,15, and developmental toxicity16. The application to aquatic toxicology has also been demonstrated, with focus on bioaccumulation and biotransformation17.
The QSAR Toolbox has been proven useful in predicting the short-term toxicity of a broad range of chemicals17, as well as the estrogen receptor (ER) binding affinities of EDs18. However, the acute ecotoxicities of EDs in aquatic vertebrates has not been analyzed using the QSAR Toolbox. In this study, a typical and facile protocol is presented to perform QSAR modeling on the acute adverse effects of EDs with a focus in fish species. The study shows that the QSAR Toolbox is a highly accessible software for calculating and predicting the lethality/mortality of aquatic vertebrates for some representative EDs. Statistical treatment methods for the derived in silico datasets are presented. Figure 1 shows the overall scheme for the general operation of the QSAR Toolbox. The workflow shown in Figure 2 provides straightforward instructions on how to operate the in silico assay to predict acute ecotoxicity of target substances such as endocrine disrupting chemicals.
Protocol
1. Equipment
- Software: use OECD QSAR Toolbox 4.0 or newer (free download from <https://qsartoolbox.org/download/?) and data analysis software.
- Computer: for the OECD QSAR Toolbox, use: (i) system type: 64 bit, Windows 7 or newer; (ii) processor: I5 at 2.4 GHz, or a faster processor or equivalent AMD CPU; (iii) installed memory (RAM): 6 GB; (iv) hard disk drive (HDD): 20 GB of free hard drive space (OECD QSAR Toolbox 4.3 Release Notes: <https://qsartoolbox.org/file/2019/02/Toolbox-4.3-Release-Notes-1.pdf>).
2. Procedure
- OECD QSAR Toolbox
NOTE: The QSAR Toolbox operates in six consecutive flow modules starting from Input and followed by Profiling, Data, Category Definition, Data Gap Filling, then Report, located at the top of the program interface.- Explore the aforementioned six stages through six toolbar icons by left-clicking. First, look over the stages of Input, Data Gap Filling, and Report that are necessary to perform the automated workflow “Ecotoxicological endpoint” and to document its results.
- Take a short look over optional stages Profiling and Data. The Profiling stage provides an initial insight into the target substance’s (eco)toxicity potential and environmental fate characteristics. Optional Data stage enables searching for available experimental data related to the target substance.
- Input
- Upon starting the QSAR Toolbox, the user begins at the Input toolbox stage by default. The QSAR Toolbox creates a working file named “Document 1” automatically, which is displayed in the stage option panel on the left of the program interface. Rename the file, if desired, by right-clicking the working file.
- Click on the CAS# button in the actions toolbar, enter the chemical abstract service (CAS) number of the target substance in the available text field, and click Search. The tool will then search for the target substance by CAS number.
- If required, choose other search options that are available in the action toolbar such as searching by substance name or simplified molecular-input line-entry system (SMILES) code. SMILES can be entered as 2D non-stereochemical or 3D stereochemical containing forms. Click Name or Structure, respectively. Use the Structure tool to draw the target substance.
- The search tool displays the search results through database records in a pop-up window. Choose the record reporting a “high” CAS-SMILES relation (CS Relation field) if multiple records are retrieved for the target substance by checking the box on the left of the record. Click OK.
NOTE: Proceeding from this point is possible only if the retrieved record contains a SMILES code, as the SMILES code (2D non-stereochemical containing form) is the basis for computation. - Batch mode: to perform the in silico assay for multiple target substances, write a simple substance list in a text editor in which each CAS number is listed in a single row (Supplementary Figure S3). Save the text file with an appropriate name and extension .txt on the computer.
- Batch mode: click Data. Then, go to Databases in the stage option panel on the left of the program interface. Make sure databases that are listed under Ecotoxicological Information are checked.
- Batch mode: click Input. Select Query from the actions toolbar. Accept the settings set in step 2.2.6 by clicking Yes in the dialog window.
- Batch mode: choose the CAS tab. Upload the substance list saved as text file through Load list from your computer.
- Batch mode: there are two Add buttons available; click the Add button on the bottom of the pop-up menu and then click Execute. The QSAR Toolbox will display a message on the number of substances that have been retrieved for the search.
NOTE: Some substances of the loaded list may not be found by the search tool or that several entries may be available for one CAS number. It is not possible to delete substances from the retrieved set of substances.
- Profiling
NOTE: The following section is optional. If this is not required, skip to section 2.5.- Click on the toolbox stage button Profiling. Go to Profiling methods in the stage option panel on the left of the program interface.
- Click Unselect All. Check all profilers listed under Predefined and those related to aquatic toxicity listed under Endpoint specific such as “Acute aquatic toxicity classification by Verhaar (Modified).”
- Finish the selection. Then click on the Apply button in the Actions toolbar.
NOTE: The QSAR Toolbox provides recommendations on a set of profilers. These are highlighted in green (suitable) and orange (plausible) when choosing Options > Color by: > Endpoint selected in the data matrix in the upper left corner of Profiling methods. Left-click the data matrix field next to the endpoint of interest. Available endpoints are listed in the endpoint tree next to the stage option panel. The profiler Substance type will indicate whether the target substance is a “discrete chemical.” The information is displayed in the expanded endpoint tree “Profile”, “Predefined”, and “Substance type”. Only if the target substance is a discrete chemical can the automated workflow run successfully. “Acute aquatic toxicity classification by Verhaar (modified)” provides a first estimate of the acute aquatic toxicity mechanism of the target substance19,20. The information is displayed in the expanded endpoint tree “Profile”, “Endpoint Specific”, and “Acute aquatic toxicity classification by Verhaar (modified)”. Five classes are available: (class 1) inert chemicals (baseline toxicity); (class 2) less inert chemicals; (class 3) reactive chemicals; (class 4) specifically acting chemicals; and (class 5) for chemicals not possible to classify. - Right-click Parameter in the endpoint tree to run integrated 2D and 3D QSAR models available in the QSAR Toolbox, if desired. Click Calculate/extract all parameters for all chemicals in the pop-up menu.
- 2D and 3D QSAR models compiled in Parameter provide numeric values. Use “Profiling methods” for qualitative information (see step 2.3.1).
- Data
NOTE: This section is optional. If it is not required, skip to section 2.5.- Click on the toolbox stage button Data. Then, click Gather from the Actions toolbar.
- Select All endpoints to gather all experimental data, then Choose to gather endpoint specific experimental data. As an example, if aquatic toxicity is the user’s focus, click Choose > Ecotoxicological Information > Aquatic toxicity > OK.
NOTE: Choosing to gather experimental data for all endpoints may lead to extended processing time. The user can adapt the hierarchy of the endpoint tree to the specific purpose. This changes the manner in which data are displayed. - If desired, right-click the endpoint of interest in the endpoint tree area. Choose Set tree hierarchy in the pop-up menu. Organize the endpoint tree in the preferred manner using the available terms and arrows and click OK.
- If desired, export the gathered data as an Excel file. Right-click on the endpoint of interest and choose Export Data matrix in the pop-up menu.
- A “Matrix export” wizard opens and enables adding other endpoints to the export list. Finish the selection, click Export, and save the file on the computer.
NOTE: Exporting data from all databases is not possible. For example, data retrieved from the database “ECHA CHEM” cannot be saved.
- Data gap filling
- Click on the toolbox stage button Data gap filling. Then, click Automated in the Actions toolbar.
- Select Ecotoxicological Endpoint > Fish, LC50 (lethal concentration, 50%) at 96 h for Pimephales promelas (mortality). Click OK. A “Workflow Controller” will appear, and processing will take up to several minutes, especially in batch mode.
NOTE: The QSAR Toolbox automatically applies a defined set of profilers when searching for suitable substances with available experimental data for the prediction. The experimental data [e.g., effect concentrations 96 h LC50 (P. promelas) or 96 h EC50 (P. promelas, mortality)] are used to generate the prediction for the target substance by either linear approximation or nearest neighbor method. Note that the methods of linear approximation and nearest neighbor are referred to as trend analysis (labeled as “T”) and read-across (labeled as “R”), respectively. - The user will receive a message if the prediction is performed successfully. Click OK and close the workflow controller indicating “Finished workflow” by clicking x in the upper right corner.
- Batch mode: upon starting the automated workflow, the user will be asked to specify the range of substances over which to execute the workflow. Accept the full range of substances selected by default in the dialog window by clicking OK.
- Batch mode: the user will not receive a message indicating whether a prediction was run successfully or unsuccessfully. Close the workflow controller indicating “Finished workflow” at the end of the batch processing by clicking x in the upper right corner.
- Report
- If a prediction was successfully executed, click on the toolbox stage button Report.
NOTE: No reports can be generated in batch mode. - Scroll down and find the prediction value in matrix field located in a yellow highlighted row next to endpoint “96-h.” The predicted value is labelled with “T” or “R.” Activate this specific data matrix field by left-clicking it.
- Click Prediction in the Actions toolbar. Customize the report content and appearance in the pop-up wizard. Three types of reports are available: (i) prediction, (ii) category, and (iii) data matrix.
- The wizard allows the user to fill in the author’s name and contact details. Write a short summary, provide a detailed explanation of the mechanistic interpretation, or provide justification for the adequacy of the prediction.
- Include additional information related to the executed prediction, if desired. The extent of additional information depends on the user.
- Go through the wizard by clicking Next. Finally, click Create report and save the prediction and category reports as PDF files and the data matrix as an Excel spreadsheet on the computer.
- Find additional details on the functionalities of the QSAR Toolbox and automated workflows in the application manual for the OECD QSAR Toolbox v.4 (F1 help on the keyboard). Details on the algorithms and rationale behind the automated workflow are described by Dimitrov et al.8 and Yordanova et al.9.
- If a prediction was successfully executed, click on the toolbox stage button Report.
3. Application
- If using the predicted effect concentration (i.e., 96-h LC50 of P. promelas) in the environmental risk assessment, use the lower limit of the 95% confidence interval. Find the data on the first page of the saved prediction report (PDF) in “Prediction summary”, “Predicted value: <mean> (from <lower_limit> to <upper_limit>).”
NOTE: The notes given here are based on results of the comparison between predicted and experimental data for a set of target substances reported in this study. Selecting the lower end of the 95% confidence range will increase the likelihood that the predicted effect concentration will not underestimate the real toxicity of the substance (see the representative results). The predicted effective concentration of the lower limit of the 95% confidence interval will therefore present a safer basis for risk assessment.
Representative Results
The example described in this study was implemented for quantitative analysis and prediction of acute toxicities of selected EDs in fish. When the predicted data points were plotted versus experimental data points as a log-log scale, a positive correlation between both was found for all fish and a representative species, namely, Pimephales promelas (fathead minnow; Figure 3). In both cases, the slope of the linear regression appeared to be comparable (predicted LC50/experimental LC50 = 0.611 and 0.602 for all fish and P. promelas, respectively). Because of the limited amount of experimental data, the number of available values from experimental observation was usually smaller than that from computational prediction. Application of the tolerance factor as 5-fold for the computational capability21 resulted in 94% (34/36) and 96% (26/27) of the protective prediction for all fish and P. promelas, respectively. Based on this prediction, 3',5,7-trihydroxy-4',6-dimethoxyisoflavone and 1,4-benzenediol appeared to exhibit calculated LC50 values greater than the tolerance limit.
To enable safety assessment at the highest reliability, further computational analysis was performed by plotting the predicted lower limit of the 95% confidence interval of LC50 (instead of the mean values used in Figure 3) versus the experimentally derived values (Figure 4). In this evaluation with an elevated safety threshold, 92% (33/36) of the total tested endocrine disrupting compounds were shown to fall into the protective range when compared to the experimentally derived values except for: 3',5,7-trihydroxy-4',6-dimethoxyisoflavone; 1,4-benzenediol; and 4-hexylphenol.
Based on assessments of the entire species available from the database, values for the predicted and experimental 96-h log10LC50 exhibited linearity with the log10KOW values in the domain between -1 and 7, indicating a hyperbolic correlation between LC50 and KOW. An overall trend existed whereby the LC50 decreased for higher KOW values of EDs for the data obtained from both computational predictions and experiments, suggesting increasing acute toxicity in fish species for EDs with higher hydrophobicity (Supplementary Figure S1).
By the rule-based ER profiler embedded in the OECD QSAR Toolbox, the ER binding affinities of the EDs were categorized as non-binding as well as weak, moderate, strong, and very strong binders, in order of increasing binding affinity18. Accordingly, the statistical distribution of log10Kow could be displayed as a qualitative classification of ER binding affinity (Supplementary Figure S2). Overall, the changes in Kow distribution ranges and their mean levels appeared to not have a defined tendency. Similarly, the distributions of predicted and experimental LC50 were shown as the extent of ER binding affinity (Figure 5). In this case, mean levels of predicted LC50 for ER binders were higher than those of non-binders. By contrast, for the experimental LC50, the mean levels of non- and weak binders were higher than those of stronger ER binders.
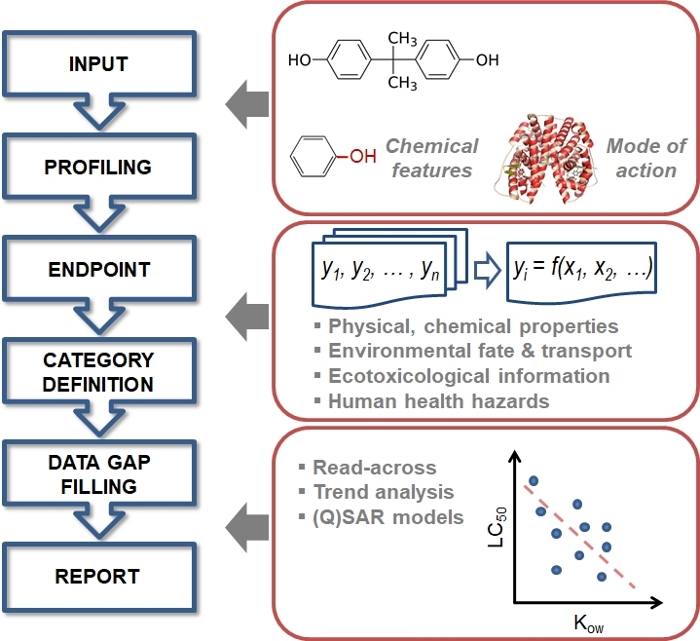
Figure 1: Basic scheme of the general workflow of the OECD QSAR Toolbox.
Please click here to view a larger version of this figure.
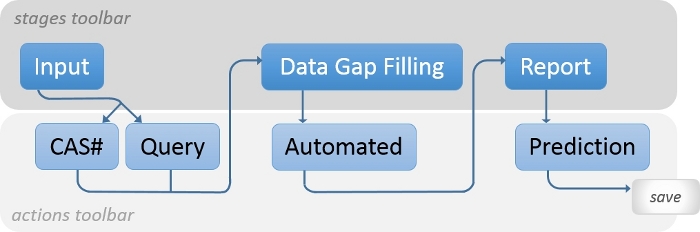
Figure 2: Workflow.
Shown is the workflow conceptualizing the modules and sequences applied to predict the acute toxicities of endocrine disruptors (EDs) in fish using the OECD QSAR Toolbox. Please click here to view a larger version of this figure.
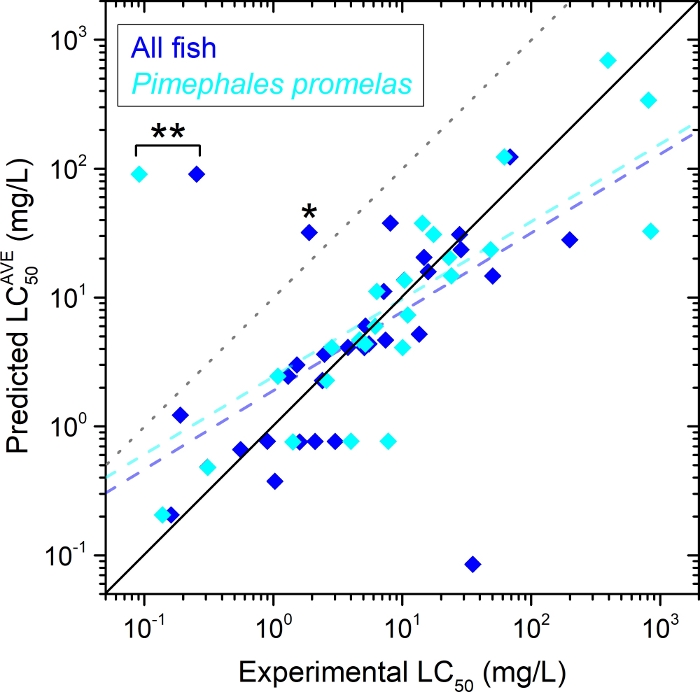
Figure 3: Predicted vs. experimental 96-h LC50 of EDs in Table 1 for all fish (blue diamonds, n = 36) and a selected species P. promelas (cyan diamonds, n = 27).
For the predicted LC50, the average (“AVE”) values are displayed. The dashed lines represent linear regressions for the two groups: for all fish (light blue), predicted LC50AVE = 0.611 x (experimental LC50) + 0.277 (adjusted r2 = 0.408); and for P. promelas (light cyan), predicted LC50AVE = 0.602 x (experimental LC50) + 0.385 (adjusted r2 = 0.441). The solid diagonal line shows unity in which the predicted and experimental values are equal21. The dotted gray line shows the 5-fold tolerance limit of the computational capability19. Outliers: 3',5,7-trihydroxy-4',6-dimethoxyisoflavone (*) and 1,4-benzenediol (**). Please click here to view a larger version of this figure.
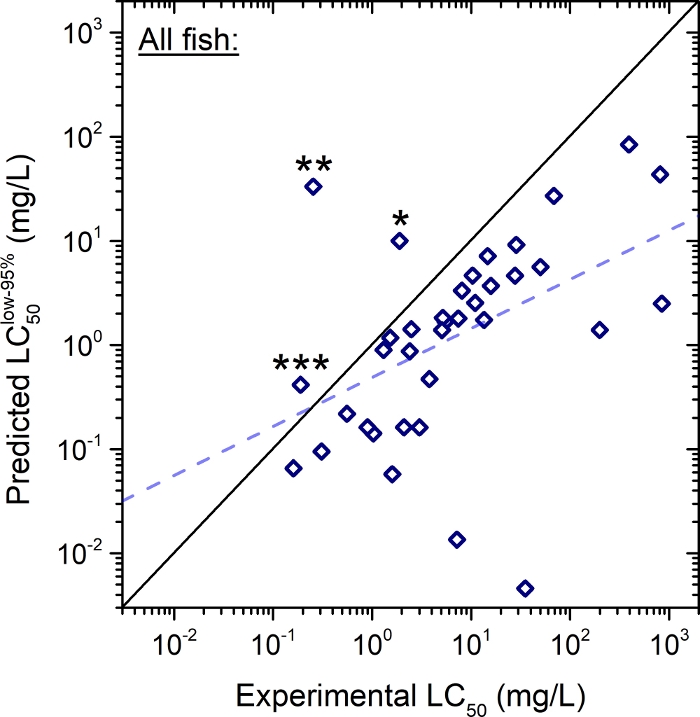
Figure 4: Predicted (lower limit of 95% confidence interval, “low-95%”) vs. experimental 96-h LC50 of EDs in Table 1 for all fish (n = 36).
The dashed line represents the linear regression: predicted LC50low-95% = 0.470 x (experimental LC50) - 0.312, where adjusted r2 = 0.193. The solid diagonal line indicates unity where the predicted and experimental values are equal to each other19. Outliers: 3',5,7-trihydroxy-4',6-dimethoxyisoflavone (*), 1,4-benzenediol (**), and 4-hexylphenol (***). Please click here to view a larger version of this figure.
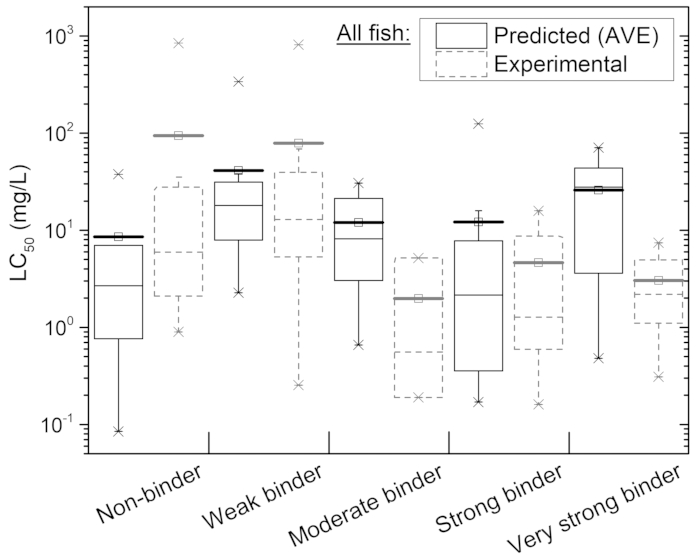
Figure 5: Distributions of predicted (solid boxes, n = 8–20 for each category) and experimental (dashed boxes; n = 3–16 for each category) 96-h LC50 depending on ER binding affinity of EDs in Table 1 for all fish.
A box plot represents: (A) mean (small square with a bold horizontal bar), (B) 1st and 3rd quartiles (lower and upper– ends of the box, respectively), (C) median (horizontal segment inside the box), (D) 5th and 95th percentile (lower and upper error bars, respectively), (E) 1st and 99th percentile (lower and upper x, respectively), and (F) minimum and maximum (lower and upper –, respectively). Please click here to view a larger version of this figure.
| No. | CAS Registry Number | Substance Name | SMILES Formula (2D non-stereochemical form) | Log Kow | AVE predicted 96-h LC50 (mg/L) | LOWER 95% CI predicted 96-h LC50 (mg/L) | Profiler - Estrogen Receptor Binding | |||||||
| 1 | 50-28-2 | 17-β Estradiol | CC12CCC3C(CCc4cc(O)ccc34)C1CCC2O | 4.01 | 3.62 | 1.42 | Very strong binder, OH group | |||||||
| 2 | 57-63-6 | 17-α Ethinyl- estradiol | CC12CCC3C(CCc4cc(O)ccc34)C1CCC2(O)C#C | 3.67 | 3.00 | 1.18 | Strong binder, OH group | |||||||
| 3 | 80-05-7 | 2,2-bis(4-hydroxyphe-nyl)propane (Bisphenol A) | CC(C)(c1ccc(O)cc1)c1ccc(O)cc1 | 3.32 | 4.68 | 1.80 | Very strong binder, OH group | |||||||
| 4 | 80-46-6 | 4-tert-Pentylphenol | CCC(C)(C)c1ccc(O)cc1 | 3.91 | 2.27 | 0.87 | Weak binder, OH group | |||||||
| 5 | 140-66-9 | 4-tert-Octylphenol | CC(C)(C)CC(C)(C)c1ccc(O)cc1 | 5.28 | 0.38 | 0.14 | Strong binder, OH group | |||||||
| 6 | 446-72-0 | Genistein [3',5,7-trihydroxy-4',6-dimethoxyisoflavone] | Oc1ccc(cc1)C1=COc2cc(O)cc(O)c2C1=O | 2.84 | 32.00 | 10.03 | Very strong binder, OH group | |||||||
| 7 | 10161-33-8 | 17β-Trenbolone | CC12C=CC3C(CCC4=CC(=O)CCC=34)C1CCC2O | 2.65 | 124.72 | 19.75 | Strong binder, OH group | |||||||
| 8 | 67747-09-5 | Prochloraz (DMI fungicide) | CCCN(CCOc1c(Cl)cc(Cl)cc1Cl)C(=O)n1ccnc1 | 4.1 | 5.19 | 1.74 | Non binder, without OH or NH2 group | |||||||
| 9 | 84852-15-3 | 4-Nonylphenol | CC(C)CCCCCCc1ccc(O)cc1 | 5.92 | 0.21 | 0.07 | Strong binder, OH group | |||||||
| 10 | 69-72-7 | salicylic acid | OC(=O)c1ccccc1O | 2.26 | 24.07 | 9.31 | Weak binder, OH group | |||||||
| 11 | 80-09-1 | 4,4’-dihydroxydiphenyl sulphone (Bisfenol S) | Oc1ccc(cc1)S(=O)(=O)c1ccc(O)cc1 | 1.65 | 48.67 | 10.67 | Very strong binder, OH group | |||||||
| 12 | 84-74-2 | phthalic acid, dibutyl ester | CCCCOC(=O)c1ccccc1C(=O)OCCCC | 4.5 | 0.76 | 0.06 | Non binder, without OH or NH2 group | |||||||
| 13 | 92-88-6 | 4,4′-dihydroxybiphenyl | Oc1ccc(cc1)-c1ccc(O)cc1 | 2.8 | 12.05 | 4.20 | Moderate binder, OH grooup | |||||||
| 14 | 94-13-3 | 4-hydroxybenzoic acid, propyl ester | CCCOC(=O)c1ccc(O)cc1 | 3.04 | 10.32 | 3.86 | Moderate binder, OH grooup | |||||||
| 15 | 98-54-4 | 4-tert-butylphenol | CC(C)(C)c1ccc(O)cc1 | 3.31 | 4.36 | 1.68 | Weak binder, OH group | |||||||
| 16 | 97-23-4 | 2,2′-dihydroxy-–5,5′-dichlorodiphenyl-methane | Oc1ccc(Cl)cc1Cc1cc(Cl)ccc1O | 4.26 | 0.48 | 0.10 | Very strong binder, OH group | |||||||
| 17 | 97-53-0 | eugenol | COc1cc(CC=C)ccc1O | 2.27 | 14.70 | 5.60 | Weak binder, OH group | |||||||
| 18 | 99-76-3 | 4-hydroxybenzoic acid, methyl ester | COC(=O)c1ccc(O)cc1 | 1.96 | 38.20 | 14.01 | Weak binder, OH group | |||||||
| 19 | 103-90-2 | N-(4-hydroxyphenyl) acetamide | CC(=O)Nc1ccc(O)cc1 | 0.46 | 338.97 | 43.39 | Weak binder, OH group | |||||||
| 20 | 106-44-5 | p-cresol | Cc1ccc(O)cc1 | 1.94 | 20.47 | 7.14 | Weak binder, OH group | |||||||
| 21 | 108-39-4 | m-cresol | Cc1cccc(O)c1 | 1.96 | 23.45 | 9.17 | Weak binder, OH group | |||||||
| 22 | 108-45-2 | 1,3-phenylenediamine | Nc1cccc(N)c1 | -0.33 | 34.60 | 0.00 | Weak binder, NH2 group | |||||||
| 23 | 108-46-3 | 1,3-dihydroxybenzene | Oc1cccc(O)c1 | 0.8 | 123.03 | 27.06 | Weak binder, OH group | |||||||
| 24 | 108-91-8 | cyclohexylamine | NC1CCCCC1 | 1.49 | 28.08 | 1.40 | Weak binder, NH2 group | |||||||
| 25 | 119-36-8 | salicylic acid, methyl ester | COC(=O)c1ccccc1O | 2.55 | 16.16 | 5.68 | Weak binder, OH group | |||||||
| 26 | 120-47-8 | 4-hydroxybenzoic acid, ethyl ester | CCOC(=O)c1ccc(O)cc1 | 2.47 | 19.93 | 7.40 | Weak binder, OH group | |||||||
| 27 | 120-80-9 | 1,2-dihydroxybenzene | Oc1ccccc1O | 0.88 | 11.14 | 0.01 | Weak binder, OH group | |||||||
| 28 | 123-31-9 | 1,4-dihydroxybenzene [1,4-benzenediol] | Oc1ccc(O)cc1 | 0.59 | 90.75 | 33.19 | Weak binder, OH group | |||||||
| 29 | 131-53-3 | 2,2′-dihydroxy-4-methoxybenzophenone | COc1ccc(C(=O)c2ccccc2O)c(O)c1 | 3.82 | 3.97 | 1.46 | Very strong binder, OH group | |||||||
| 30 | 131-56-6 | 2,4-dihydroxybenzophenone | Oc1ccc(c(O)c1)C(=O)c1ccccc1 | 2.96 | 12.04 | 4.73 | Strong binder, OH group | |||||||
| 31 | 131-57-7 | 2-hydroxy-4-methoxybenzophenone | COc1ccc(C(=O)c2ccccc2)c(O)c1 | 3.79 | 5.96 | 2.27 | Strong binder, OH group | |||||||
| 32 | 599-64-4 | 4-cumylphenol | CC(C)(c1ccccc1)c1ccc(O)cc1 | 4.12 | 2.15 | 0.84 | Strong binder, OH group | |||||||
| 33 | 2855-13-2 | 1-amino-3-aminomethyl-3,5,5-trimethyl-cyclohexane | CC1(C)CC(N)CC(C)(CN)C1 | 1.9 | 30.65 | 1.53 | Moderate binder, NH2 group | |||||||
| 34 | 6864-37-5 | 3,3′-dimethyl-4,4′-diaminodicyclohexylmethane | CC1CC(CCC1N)CC1CCC(N)C(C)C1 | 4.1 | 1.07 | 0.05 | Strong binder, NH2 group | |||||||
| 35 | 25013-16-5 | tert-butyl-4-hydroxyanisole | COc1ccc(O)c(c1)C(C)(C)C | 3.5 | 4.85 | 1.85 | Moderate binder, OH grooup | |||||||
| 36 | 147315-50-2 | 2-(4,6-diphenyl-1,3,5-triazin-2-yl)-5-(hexyloxy)phenol | CCCCCCOc1ccc(c(O)c1)-c1nc(nc(n1)-c1ccccc1)-c1ccccc1 | 6.24 | 0.17 | 0.06 | Strong binder, OH group | |||||||
| 37 | 88-68-6 | 2-aminobenzamide | NC(=O)c1ccccc1N | 0.35 | 694.00 | 84.30 | Weak binder, NH2 group | |||||||
| 38 | 611-99-4 | 4,4′-dihydroxybenzophenone | Oc1ccc(cc1)C(=O)c1ccc(O)cc1 | 2.19 | 37.74 | 14.67 | Very strong binder, OH group | |||||||
| 39 | 27955-94-8 | 1,1,1-tris(4-hydroxyphenol)ethane | CC(c1ccc(O)cc1)(c1ccc(O)cc1)c1ccc(O)cc1 | 4.38 | 2.09 | 0.82 | Very strong binder, OH group | |||||||
| 40 | 87-18-3 | salicylic acid, 4-tert-butylphenyl ester | CC(C)(C)c1ccc(OC(=O)c2ccccc2O)cc1 | 5.73 | 0.24 | 0.09 | Strong binder, OH group | |||||||
| 41 | 47465-97-4 | 3,3-bis(3-methyl-4-hydroxyphenyl)2-indolinone | Cc1cc(ccc1O)C1(C(=O)Nc2ccccc12)c1ccc(O)c(C)c1 | 4.48 | 2.07 | 0.77 | Very strong binder, OH group | |||||||
| 42 | 99-96-7 | p-hydroxybenzoic acid | OC(=O)c1ccc(O)cc1 | 1.58 | 8.54 | 0.00 | Weak binder, OH group | |||||||
| 43 | 80-07-9 | 1-Chloro-4-(4- chlorophenyl)sulfonylbenz | Clc1ccc(cc1)S(=O)(=O)c1ccc(Cl)cc1 | 3.9 | 3.92 | 0.85 | Non binder, without OH or NH2 group | |||||||
| 44 | 84-65-1 | 9,10-Anthraquinone | O=C1c2ccccc2C(=O)c2ccccc12 | 3.39 | 7.00 | 3.54 | Non binder, without OH or NH2 group | |||||||
| 45 | 85-44-9 | 2-benzofuran-1,3-dione | O=C1OC(=O)c2ccccc12 | 1.6 | 2.69 | 0.00 | Non binder, without OH or NH2 group | |||||||
| 46 | 92-84-2 | 10H-Phenothiazine | N1c2ccccc2Sc2ccccc12 | 4.15 | 1.07 | 0.08 | Non binder, without OH or NH2 group | |||||||
| 47 | 2855-13-2 | 1-amino-3-aminomethyl-3,5,5-trimethyl-cyclohexane | CC1(C)CC(N)CC(C)(CN)C1 | 1.9 | 30.65 | 1.53 | Moderate binder, NH2 group | |||||||
| 48 | 50-27-1 | Estriol | CC12CCC3C(CCc4cc(O)ccc34)C1CC(O)C2O | 2.45 | 21.21 | 8.29 | Very strong binder, OH group | |||||||
| 49 | 50-50-0 | beta-Estradiol-3-benzoate | CC12CCC3C(CCc4cc(OC(=O)c5ccccc5)ccc34)C1CCC2O | 5.47 | 0.36 | 0.02 | Strong binder, OH group | |||||||
| 50 | 53-16-7 | Estrone | CC12CCC3C(CCc4cc(O)ccc34)C1CCC2=O | 3.13 | 7.78 | 3.06 | Strong binder, OH group | |||||||
| 51 | 92-52-4 | Biphenyl | c1ccc(cc1)-c1ccccc1 | 4.01 | 4.10 | 0.47 | Non binder, without OH or NH2 group | |||||||
| 52 | 92-69-3 | p-Phenylphenol | Oc1ccc(cc1)-c1ccccc1 | 3.2 | 5.99 | 1.82 | Moderate binder, OH grooup | |||||||
| 53 | 96-29-7 | 2-Butanone oxime | CCC(C)=NO | 0.63 | 32.67 | 2.49 | Non binder, non cyclic structure | |||||||
| 54 | 121-75-5 | Malathon | CCOC(=O)CC(SP(=S)(OC)OC)C(=O)OCC | 2.36 | 37.73 | 3.33 | Non binder, non cyclic structure | |||||||
| 55 | 123-07-9 | 4-Ethylphenol | CCc1ccc(O)cc1 | 2.58 | 13.63 | 4.65 | Weak binder, OH group | |||||||
| 56 | 645-56-7 | 4-n-Propylpehnol | CCCc1ccc(O)cc1 | 3.2 | 7.32 | 2.55 | Weak binder, OH group | |||||||
| 57 | 1638-22-8 | p-Butyl phenol | CCCCc1ccc(O)cc1 | 3.65 | 4.09 | 1.39 | Weak binder, OH group | |||||||
| 58 | 1912-24-9 | Atrazine | CCNc1nc(Cl)nc(NC(C)C)n1 | 2.61 | 30.87 | 4.63 | Non binder, without OH or NH2 group | |||||||
| 59 | 40596-69-8 | Methoprene | COC(C)(C)CCCC(C)CC=CC(C)=CC(=O)OC(C)C | 5.5 | 0.08 | 0.00 | Non binder, non cyclic structure | |||||||
| 60 | 1987-50-4 | 4-Heptylphenol | CCCCCCCc1ccc(O)cc1 | 5.01 | 0.66 | 0.22 | Moderate binder, OH grooup | |||||||
| 61 | 92-86-4 | p,p'-Dibromobiphenyl | Brc1ccc(cc1)-c1ccc(Br)cc1 | 5.72 | 0.11 | 0.02 | Non binder, without OH or NH2 group | |||||||
| 62 | 480-41-1 | Naringenin | Oc1ccc(cc1)C1CC(=O)c2c(O)cc(O)cc2O1 | 2.52 | 27.84 | 10.87 | Very strong binder, OH group | |||||||
| 63 | 486-66-8 | Daidzein | Oc1ccc(cc1)C1=COc2cc(O)ccc2C1=O | 2.55 | 36.47 | 11.71 | Very strong binder, OH group | |||||||
| 64 | 491-70-3 | Luteolin | Oc1cc(O)c2C(=O)C=C(Oc2c1)c1ccc(O)c(O)c1 | 2.53 | 43.75 | 14.28 | Very strong binder, OH group | |||||||
| 65 | 491-80-5 | Biochanin A | COc1ccc(cc1)C1=COc2cc(O)cc(O)c2C1=O | 3.41 | 15.87 | 3.70 | Strong binder, OH group | |||||||
| 66 | 520-18-3 | Kaempferol | Oc1ccc(cc1)C1Oc2cc(O)cc(O)c2C(=O)C=1O | 1.96 | 70.98 | 8.05 | Very strong binder, OH group | |||||||
| 67 | 2051-60-7 | 2-Chlorobiphenyl (PCB 1) | Clc1ccccc1-c1ccccc1 | 4.53 | 0.77 | 0.16 | Non binder, without OH or NH2 group | |||||||
| 68 | 2051-61-8 | 3-Chlorobiphenyl (PCB 2) | Clc1cccc(c1)-c1ccccc1 | 4.58 | 0.77 | 0.16 | Non binder, without OH or NH2 group | |||||||
| 69 | 2051-62-9 | 4-Chloro-1,1'-biphenyl | Clc1ccc(cc1)-c1ccccc1 | 4.61 | 0.77 | 0.16 | Non binder, without OH or NH2 group | |||||||
| 70 | 2446-69-7 | p-n-Hexylphenol [4-hexylphenol] | CCCCCCc1ccc(O)cc1 | 4.52 | 1.22 | 0.42 | Moderate binder, OH grooup | |||||||
| 71 | 14938-35-3 | 4-n-Amylphenol | CCCCCc1ccc(O)cc1 | 4.06 | 2.44 | 0.89 | Weak binder, OH group | |||||||
| 72 | 17924-92-4 | Zearalenone | CC1CCCC(=O)CCCC=Cc2cc(O)cc(O)c2C(=O)O1 | 3.58 | 7.22 | 2.66 | Strong binder, OH group | |||||||
| 73 | 1743-60-8 | beta-Estradiol 3-benzoate 17-nbutyrate | CC(=O)OC1CCC2C3CCc4cc(O)ccc4C3CCC12C | 4.95 | 0.91 | 0.35 | Strong binder, OH group | |||||||
| 74 | 479-13-0 | Coumestrol | Oc1ccc2c(OC(=O)c3c-2oc2cc(O)ccc32)c1 | 1.57 | 52.16 | 11.44 | Very strong binder, OH group | |||||||
Table 1: List of evaluated endocrine disrupting chemicals. Average mean (AVE) and lower 95% confidence interval (CI) effective concentrations (95-h LC50, Pimephales promelas) as well as Estrogen Receptor Binding were predicted with the QSAR Toolbox version 4.3 Automated Workflow. Log10Kow was retrieved via QSAR Toolbox version 4.3 from KOWWIN v1.68, 2000, U.S. Environmental Protection Agency. Experimental log10Kow values were preferred over predicted values. The target substance list was compiled from previously reported lists of EDs22,23,24.
Supplementary Information. Please click here to download this file.
Discussion
The versatility of the OECD QSAR Toolbox as analytic software for ecotoxicology is shown here with specific interest in the adverse effects of endocrine disrupting chemicals on aquatic vertebrates. In addition, a simple and standard protocol was demonstrated for predicting acute toxicity (96-h LC50) of 74 representative EDs (Table 1) for fish species. This was achieved by applying category building, data gap filling, and ER profiling modules embedded in the QSAR Toolbox (Figure 1, Figure 2).
The linear correlation between log10LC50 and log10KOW with a negative slope (as shown in Supplementary Figure S1) has long been known as a standard quantitative relationship in QSAR analyses25, where higher toxicity is shown the more hydrophobic a given chemical is. As can be seen from a simple calculation, the general mathematical relation that includes Equation S1 and Equation S2 (Supplementary Information) is a converted expression from the following power function26:
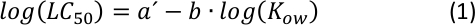

From the plot of (Equation 2), characterizing an intermediate range of KOW26 may be possible by adjusting the parameters a and b, where a certain variation in hydrophobicity (or hydrophilicity) does not significantly change the endpoint of acute toxicity.
Comparative analyses between the computational predictions and experimental observations on the LC50, as shown in Figure 3 and Figure 4, have been typically reported in studies of QSAR for various aquatic toxicants, including technical nonionic surfactants27, triazole fungicides28, and pesticide metabolites21. This type of retrospective validation provides information on how far a given QSAR tool can reach in terms of comparative performance to experimental results. In this study of acute toxicity in fish, the QSAR Toolbox was proven to provide protective predictions for over 90% of tested EDs in all fish and in a single species, Pimephales promelas.
Further identifying the three outlier chemicals in Figure 3 and Figure 4, which showed higher predicted LC50 on average and at a minimum, respectively, is required. First, the 3',5,7-trihydroxy-4',6-dimethoxyisoflavone is a type of flavonoid (more specifically, an isoflavone), which is considered to be generally safe and used in herbal pharmaceuticals; however, it still has estrogen-related concerns29 and may cause acute toxicity probably through oxidative phosphorylation uncoupling30. Next, the 1,4-benzenediol, called hydroquinone, is a phenolic compound that can trigger a non-specific and cytotoxic immune response in fish31. Finally, the 4-hexylphenol has been known to exhibit sufficient positive estrogenic activity to be classified as an ED32. It has been well-studied that the main reason of the acute toxicity of hydroquinone is the reduction-oxidation (redox) cycling. The hydroquinone is oxidized to benzoquinone and reduced back to semi-quinone or hydroquinone repeatedly, with depleting cofactors and generating reactive oxygen species33. The other two chemicals may require deeper investigations to reveal their mechanisms of action in acute ecotoxicity using molecular docking approaches such as that used by Panche et al.34, which cannot be covered by the QSAR Toolbox.
EDs interfere with the endocrine system mainly through physicochemical interactions with steroid receptors such as estrogen and androgen receptors, which are of considerable interest in QSAR modeling studies35. Considering this, the QSAR Toolbox is robust in terms of facile and rapid classification of ER binding affinities for a set of chemicals based only on the 2D descriptors of molecular structures. When this ER profiler system was applied to our list of EDs, no clear correlation was found between ER binding affinity and hydrophobicity (Supplementary Figure S2). This result may be explained by the fact that the formation of a steroid-receptor complex is not a direct consequence of a hydrophobic bonding contribution but should be accompanied by a conformational change in the active-site receptor structure36. The receptor binding can be also due to hydrogen-bonding and π-stacking.
Additionally, the position of each chemical group on the molecule may affect the receptor binding, even if the hydrophobicity and number of hydrogen-bond acceptors-donors remain the same. Second, the ER profiler produced contrary trends between predicted and experimental LC50 mean levels with increasing ER binding affinity (Figure 5). This may be because the lethality of parents in an acute toxicity test are not due to ER binding but rather to narcosis in most cases, or to redox cycling in the case of hydroquinone. For example, more extensive analysis, including the chronic toxicity, is required for a larger set of EDs to define predictive limitations of the current version of the QSAR Toolbox.
This preliminary research may also have public health implications because steroids (androgens, estrogens, progestines, and corticoids) and their receptors exhibit similar or even identical macromolecular structures across vertebrates5. These types of analogous endocrine signaling systems may operate using a common mechanism in key events of EDs5. Nevertheless, additional and complementary methodologies are required to illuminate this vast and complex aspect [for example, by performing computational modeling of absorption, distribution, metabolism, and excretion (ADME), and/or adverse outcome pathway (AOP)]38. Furthermore, because most of the scientific and public concerns raised about the adverse effects of EDs are related to their chronic toxicities, improving the databases and algorithms in the QSAR Toolbox and producing reliable long-term ecotoxicology predictions for EDs are both necessary.
This paper demonstrates the application of QSAR Toolbox to compare ecotoxicological LC50 values for fish with log10Kow values of EDs. Throughout the protocol, it results in weak relationships between the two parameters, as it has been revealed by previous studies (e.g., Kim et al.39) that log10Kow is not a good direct predictor of aquatic LC50. In spite of this limitation, this protocol provides a general review or “vignette” to describe how to use the dashboard for a given purpose, since it is a valid application to use the QSAR Toolbox for investigating correlations between LC50 (or ER binding affinity) and log10Kow, or as a tool for rapid acute ecotoxicity screening. Nevertheless, it should be noted that (1) illuminating the link between estrogen receptor binding and chronic toxicity, rather than acute toxicity (lethality), is more relevant so that clearer correlations may be found, and (2) the androgen receptor, together with that of estrogen, also plays a critical role in reproductive toxicity. Therefore, it is required for the future version of the QSAR Toolbox to improve the prediction functions in light of those two points.
Acknowledgements
This research was supported by the National Research Council of Science & Technology (NST) grant by the South Korean government (MSIP) (No. CAP-17-01-KIST Europe) and Project 11911.
Materials
| Name | Company | Catalog Number | Comments |
| Adobe Acrobat Reader DC | Adobe Systems Software Ireland Limited | NA | Required to view prediction and category report |
| Computer | System: Microsoft Corporation | NA | Recommended system properties: (i) system type: 64 bit, Microsoft Windows 7 or newer, (ii) processor: I5 at 2.4 GHz or faster processor or equivalent AMD CPU, (iii) Installed memory (RAM): 6 GB of RAM, (iv) Hard Disk Drive (HDD): 20 GB free hard drive space |
| Microsoft Editor | Microsoft Corporation | NA | Required to upload a substance list of CAS numbers (batch mode) to the OECD QSAR Toolbox as .txt file (text file) |
| Microsoft Excel 2016 | Microsoft Corporation | NA | Required to export data from OECD QSAR Toolbox as .cvs, .xls or .xlsx files |
| OECD QSAR Toolbox version 4.0 or newer | Organisation for Economic Co-operation and Development | NA | Required to run OECD QSAR Toolbox Automated Workflows; free download: https://qsartoolbox.org/download/ |
| OriginPro 9 | OriginLab Corporation | NA | Optional program for data analysis; similar tools possible |
References
- Najarian, K., Najarian, S., Gharibzadeh, S., Eichelberger, C. N. . Systems Biology and Bioinformatics: A Computational Approach. , (2009).
- Fujita, T., Iwasa, J., Hansch, C. A new substituent constant, π, derived from partition coefficient. Journal of the American Chemical Society. 86, 5175-5180 (1964).
- Roy, K., Kar, S., Das, R. N. . Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment. , (2015).
- Raies, A. B., Bajic, V. B. In silico toxicology: computational methods for the prediction of chemical toxicity. WIREs Computational Molecular Science. 6, 147-172 (2016).
- Hayes, T. B. Welcome to the revolution: integrative biology and assessing the impact of endocrine disruptors on environmental and public health. Integrative Compuational Biology. 45, 321-329 (2005).
- Schug, T. T., et al. Minireview: endocrine disruptors: past lessons and future directions. Molecular Endocrinology. 30, 833-847 (2016).
- Devillers, J., Marchand-Geneste, N., Carpy, A., Porcher, J. M. SAR and QSAR modeling of endocrine disruptors. SAR QSAR Environmental Research. 17, 393-412 (2006).
- Dimitrov, S. D., et al. QSAR Toolbox - workflow and major functionalities. SAR QSAR Environmental Research. 27, 203-219 (2016).
- Yordanova, D., et al. Automated and standardized workflows in the OECD QSAR Toolbox. Computational Toxicology. 10, 89-104 (2019).
- Mombelli, E., Devillers, J. Evaluation of the OECD (Q)SAR Application Toolbox and Toxtree for predicting and profiling the carcinogenic potential of chemicals. SAR QSAR Environmental Research. 21, 731-752 (2010).
- Devillers, J., Mombelli, E., Samsera, R. Structural alerts for estimating the carcinogenicity of pesticides and biocides. SAR QSAR Environmental Research. 22, 89-106 (2011).
- Li, C., et al. Identifying unknown by-products in drinking water using comprehensive two-dimensional gas chromatography-quadrupole mass spectrometry and in silico toxicity assessment. Chemosphere. 163, 535-543 (2016).
- Devillers, J., Mombelli, E. Evaluation of the OECD QSAR Application Toolbox and Toxtree for estimating the mutagenicity of chemicals. Part 1. Aromatic amines. SAR QSAR Environmental Research. 21, 753-769 (2010).
- Devillers, J., Mombelli, E. Evaluation of the OECD QSAR Application Toolbox and Toxtree for estimating the mutagenicity of chemicals. Part 2. α-β unsaturated aliphatic aldehydes. SAR QSAR Environmental Research. 21, 771-783 (2010).
- Kulkarni, S. A., Barton-Maclaren, T. S. Performance of (Q)SAR models for predicting Ames mutagenicity of aryl azo and benzidine based compounds. Journal of Environmental Science and Health Part C Environmental Carcinogenesis & Ecotoxicology Reviews. 32, 46-82 (2014).
- Craig, E. A., Wang, N. C., Zhao, Q. J. Using quantitative structure-activity relationship modeling to quantitatively predict the developmental toxicity of halogenated azole compounds. Journal of Applied Toxicology. 34, 787-794 (2014).
- Tebby, C., Mombelli, E., Pandard, P., Péry, A. R. Exploring an ecotoxicity database with the OECD (Q)SAR Toolbox and DRAGON descriptors in order to prioritise testing on algae, daphnids, and fish. Science of the Total Environment. 409, 3334-3343 (2011).
- Mombelli, E. Evaluation of the OECD (Q)SAR Application Toolbox for the profiling of estrogen receptor binding affinities. SAR QSAR Environmental Research. 23, 37-57 (2012).
- Verhaar, H. J. M., van Leeuwen, C. J., Hermens, J. L. M. Classifying environmental pollutants. 1: structure-activity relationships for prediction of aquatic toxicology. Chemosphere. 25, 471-491 (1992).
- Enoch, S. J., Hewitt, M., Cronin, M. T. D., Azam, S., Madden, J. C. Classification of chemicals according to mechanism of aquatic toxicity: an evaluation of the implementation of the Verhaar scheme in Toxtree. Chemosphere. 73, 243-248 (2008).
- Burden, N., Maynard, S. K., Weltje, L., Wheeler, J. R. The utility of QSARs in predicting acute fish toxicity of pesticide metabolites: a retrospective validation approach. Regulatory Toxicology and Pharmacology. 80, 241-246 (2016).
- Nendza, M., et al. Screening for potential endocrine disruptors in fish: evidence from structural alerts and in vitro and in vivo toxicological assays. Environmental Sciences Europe. 28, 26 (2016).
- Roncaglioni, A., Piclin, N., Pintore, M., Benfenati, E. Binary classification models for endocrine disrupter effects mediated through the estrogen receptor. SAR QSAR Environmental Research. 19, 697-733 (2008).
- Sosnovcová, J., Rucki, M., Bendová, H. Estrogen receptor binding affinity of food contact material components estimated by QSAR. Central European Journal of Public Health. 24, 241-244 (2016).
- Walker, J. D., Dearden, J. C., Schultz, T. W., Jaworska, J., Comber, M. H. I., Walker, J. D. QSARs for New Practitioners. QSARs for Pollution Prevention, Toxicity Screening, Risk Assessment, and Web Applications. , (2003).
- Sánchez-Bayo, F. From simple toxicological models to prediction of toxic effects in time). Ecotoxicology. 18, 343-354 (2009).
- Sjöström, M., Lindgren, &. #. 1. 9. 7. ;., Uppgård, L. L., Chen, F., Schüürmann, G. Joint Multivariate Quantitative Structure-Property and Structure-Activity Relationships for a Series of Technical Nonionic Surfactants. Quantitative Structure-Activity Relationships in Environmental Sciences-VII. , (1997).
- Ding, F., Guo, J., Song, W., Hu, W., Li, Z. Comparative quantitative structure-activity relationship (QSAR) study on acute toxicity of triazole fungicides to zebrafish. Chemistry Ecology. 27, 359-368 (2011).
- Galati, G., O'Brien, P. J. Potential toxicity of flavonoids and other dietary phenolics: significance for their chemopreventive and anticancer properties. Free Radical Biology in Medicine. 37, 287-303 (2004).
- Russom, C. L., Bradbury, S. P., Broderius, S. J. Predicting modes of action from chemical structure: acute toxicity in the fathead minnow (Pimephales promelas). Environmental Toxicology and Chemistry. 16, 948-967 (1997).
- Taysse, L., Troutaud, D., Khan, N. A., Deschaux, P. Structure-activity relationship of phenolic compounds (phenol, pyrocatechol and hydroquinone) on natural lymphocytotoxicity of carp (Cyprinus carpio). Toxicology. 98, 207-214 (1995).
- Nishihara, T., et al. Estrogenic activities of 517 chemicals by yeast two-hybrid assay. Journal of Health Science. 46, 282-298 (2000).
- Bolton, J. L., Trush, M. A., Penning, T. M., Dryhurst, G., Monks, T. J. Role of quinones in toxicology. Chemical Research in Toxicology. 13, 135-160 (2000).
- Panche, A. N., Diwan, A. D., Chandra, S. R. Flavonoids: an overview. Journal of Nutritional Science. 5, e47 (2016).
- Li, J., Gramatica, P. QSAR classification of estrogen receptor binders and pre-screening of potential pleiotropic EDCs. SAR QSAR Environmental Research. 21, 657-669 (2010).
- Bohl, M. . Molecular Structure and Biological Activity of Steroids. , (2017).
- Kaminuma, T., Takai-Igarashi, T., Nakano, T., Nakata, K. Modeling of signaling pathways for endocrine disruptors. BioSystems. 55, 23-31 (2000).
- Lillicrap, A., et al. Alternative approaches to vertebrate ecotoxicity tests in the 21st century: a review of developments over the last 2 decades and current status. Environmental Toxicology and Chemistry. 35, 2637-2646 (2016).
- Kim, J. W., et al. Acute toxicity of pharmaceutical and personal care products on freshwater crustacean (Thamnocephalus platyurus) and fish (Oryzias latipes). Journal of Toxicological Sciences. 34, 227-232 (2009).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved