Method Article
Quantifying Levels of Dopaminergic Neuron Morphological Alteration and Degeneration in Caenorhabditis elegans
In This Article
Summary
In this article, we showcase how to use a seven-point scoring system to consistently quantify changes to dopaminergic neuron dendrite morphology in C. elegans. This system is intended for analyses of dopaminergic neurodegeneration assays utilizing genetic, chemical, and age-based models of neurodegenerative disorders.
Abstract
Dopamine neuron loss is involved in the pathology of Parkinson's Disease (PD), a highly prevalent neurodegenerative disorder affecting over 10 million people worldwide. Since many details about PD etiology remain unknown, studies investigating genetic and environmental contributors to PD are needed to discover methods of prevention, management, and treatment. Proper characterization of dopaminergic neuronal loss may be relevant not only to PD research, but to other increasingly prevalent neurodegenerative disorders.
There are established genetic and chemical models of dopaminergic neurodegeneration in the Caenorhabditis elegans model system, with easy visualization of neurobiology supported by the nematodes' transparency and invariant neuronal architecture. In particular, hermaphroditic C. elegans' dopaminergic neuron morphological changes can be visualized using strains with fluorescent reporters driven by cell-specific promotors such as the dat-1 dopamine transporter gene, which is expressed exclusively in their eight dopaminergic neurons.
With the capabilities of this model system and the appropriate technology, many laboratories have studied dopaminergic neurodegeneration. However, there is little consistency in the way the data is analyzed and much of the present literature uses binary scoring analyses that capture the presence of degeneration but not the full details of the progression of neuron loss. Here, we introduce a universal scoring system to assess morphological changes and degeneration in C. elegans' cephalic neuron dendrites. This seven-point scale allows for analysis across a full range of dendrite morphology, ranging from healthy neurons to complete dendrite loss, and considering morphological details including kinks, branching, blebs, and breaks. With this scoring system, researchers can quantify subtle age-related changes as well as more dramatic chemical-induced changes. Finally, we provide a practice set of images with commentary that can be used to train, calibrate, and assess the scoring consistency of researchers new to this method. This should improve within- and between- laboratory consistency, increasing rigor and reproducibility.
Introduction
Parkinson's disease (PD) is an increasingly common neurodegenerative disease affecting up to 10 million individuals worldwide1. Males and older individuals are at a higher risk for developing PD; the average age of onset for the disease is 60 years, and PD incidence climbs from a 0.3% incidence in the general population to 3% in individuals over 80 years of age1,2. Although the details of PD pathology are not fully understood, this progressive disorder involves the loss of dopaminergic neurons in the substantia nigra region of the midbrain. Hypothesized mechanisms of this neuronal loss involve mitochondrial dysfunction, oxidative stress, and inflammation2. The causes and risk factors for the disease are still being explored, but involve a combination of environmental and genetic factors1. For example, studies have found positive associations between lifelong pesticide use and PD, as well as genetic susceptibility to familial PD1,3.
The C. elegans model system, originally developed in part for neurobiology research4, is well suited for evaluating dopaminergic neuron loss in vivo. Nass and colleagues pioneered the use of C. elegans for dopaminergic neurodegeneration5, and many groups have since adopted the worm as a successful model for PD and dopaminergic dysfunction6,7,8,9,10,11,12,13,14,15,16,17,18,19,20. C. elegans are good neurodegenerative disease models for many of the same reasons that they are such a popular model organism for other areas of biology; their transparency allows for in vivo study of cellular processes, genetic manipulation in worms is relatively quick and easy, they have a short generation time of about three days, and they are easy to maintain21. Most PD worm models fall into one of three categories: age-based models, chemical models, and genetic models. The ability to synchronize a population of worms allows for the study of age-related neurodegeneration for an age based-model of neurodegenerative diseases associated with aging, such as PD22. Chemical exposures inducing PD-like neuronal defects have been established using a variety of chemicals including 6-hydroxydopamine (6-OHDA), rotenone, and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)22. Worms are also successfully used as genetic models of PD; strains with select neural gene knockouts can model various neurodegenerative diseases1,4. Combinations of genetic and environmental factors, or "gene-environment interactions," which likely play a major role in PD2,17,23,24,25,26,27,28, have been examined by several groups using C. elegans. Finally, age-related dopaminergic neurodegeneration has also been observed29,30. If using an appropriate neural transgenic strain in fluorescent imaging, any of these PD worm models may be used to study dopaminergic neurodegeneration.
Quantifying changes to neuronal morphology is a critical component of neurodegenerative research. In C. elegans, many fluorescent reporter strains have been used to visualize morphological changes and loss of neurons. Strains suitable for neuronal imaging feature a fluorescent protein associated with cell-specific promoters. For dopaminergic neurodegeneration assays, our laboratory has used the BY200 [vtIs1 (dat-1p::GFP, rol-6)] strain, which has a green fluorescent protein (GFP) tag in the dat-1 gene, expressed in the dopaminergic neurons. Note that the BY200's roller phenotype has a very low penetrance and is rarely observed. Other common strains used for this type of imaging include BY250 [dat-1p::GFP], BY273 [baEx18[dat-1p::GFP+dat-1p::WT α-syn]], BZ555 [egIs1 [dat-1p::GFP]], and several others available from the Caenorhabditis Genetics Center (CGC) or upon request from specific laboratories1,21,22,29. These strains typically allow for visualization of all three classes of dopaminergic neurons: cephalic (CEP), anterior deirid (ADE), and postdeirid (PDE) neurons. C. elegans does not naturally express the alpha synuclein protein, but strains such as BY273 have been engineered to express it. However, we note that the scoring system we present was developed using BY200, which does not express alpha synuclein, and would need to be validated with that strain (or any other new strain) prior to use. Additional dopaminergic neurons are present in males but are rarely considered because males normally comprise <1% of a C. elegans population. Here, we focus on the four CEP dopaminergic neurons found in the head region of C. elegans. This set of neurons is easily located under fluorescent microscopy, is present in both hermaphrodite and male worms, does not typically overlap with other areas of auto-fluorescence, and is commonly reported on in worm studies. Notably, though these neurons are not myelinated, the CEP dorsal (CEPD) neurons are directly exposed to the pseudocoelomic body fluid where as the CEP ventral neurons are not. A healthy set of CEP dendrites typically displays as relatively straight, uninterrupted lines. Degenerated dendrites may show any combination of irregularities and signs of damage, including pronounced dots called blebs along the line of the dendrite and breaks in the line of the dendrite. Examples of CEP neurons at varying levels of degeneration can be seen in Figure 1.
Although dopaminergic neurodegeneration is being studied by a growing number of C. elegans laboratories, there has been a large variation in analytical methods of quantifying dopaminergic neuron damage29,31,32,33,34. Many published studies have reported on the presence or absence of CEP soma with a binary scoring system of degenerative versus typical or wild type neurons31,32. These scoring methods can identify certain stressors that induce neurodegeneration but cannot quantify the details of the progression of more subtle neuronal damage, or easily detect differences between neurodegeneration as induced by unique chemicals or other variables. Additionally, scoring systems focused on the cell bodies may not be sensitive to less severe levels of damage or to neuronal damage affecting only part of the cell, such as the dendrite. Since the dendrite appears to have the largest range of consistently detectable morphologic changes in response to chemical stressors, we have selected them as the basis for our analysis. The scoring system we present here is modified from dendrite morphology based multi-point scales that have been previously used in our lab29,33. This system expands these five- and six-point scales into a seven-point scale to account for age-related morphological changes, such as higher expected numbers of kinks in older adult dendrites, and to differentiate between severe damage and complete dendrite loss. The purpose of introducing this scoring system is to provide the ability to capture a comprehensive picture of neurodegeneration at all levels of neuronal damage and provide a universal system to support consistency across C. elegan dopaminergic neurodegeneration research. Because scoring is inherently subjective, it is critical to maximize consistency between individuals scoring, and to blind the scorer to the identity of the images using manual blinding or an automatic blinding program35. To improve consistency, we present a series of training images and utilize JoVE's video capabilities to demonstrate our scoring system in detail. We recommend using a system that both permits automated blinded scoring and allows the scorer to quantify her or his scoring consistency by re-scoring a subset of images. This is particularly important when combining or comparing data from multiple scientists, or training scientists new to scoring.
Protocol
1. Prepare Worms for Imaging
NOTE: See related JoVE video article31: https://www.jove.com/v/835/
- For each experimental group, pipette or pick 20 to 30 worms to an imaging platform compatible with the imaging microscope. Most common platforms include 2% agarose gel pads mounted on glass slides with a coverslip31 and 96-well plates containing well volumes at or less than 100 µL of liquid medium.
- Paralyze the worms by adding 30-90 mM sodium azide (NaN3), 2.5-8.5 mM levamisole HCl, or other paralyzing agent to the worms. If paralyzing in liquid, use a higher concentration of paralyzing agent than if paralyzing on agarose pads. Tap imaging platform gently to mix.
- Allow worms to paralyze completely.
NOTE: This may take several minutes.
2. Image Dopaminergic Neurons
- Locate worms' head regions under single-color GFP fluorescence using imaging microscope capable of taking z-stacks.
- Be mindful of exposure and aperture settings; avoid overexposure of dendrites and keep settings consistent across trials. Make the dendrites as bright as needed for clear visualization; this typically results in overexposure of the soma.
NOTE: Images included in this protocol were captured using 400x magnification.
- Be mindful of exposure and aperture settings; avoid overexposure of dendrites and keep settings consistent across trials. Make the dendrites as bright as needed for clear visualization; this typically results in overexposure of the soma.
- Scroll through the focus to find upper and lower bounds where the dendrites are clear. Set these as upper and lower bounds for a z-stack image capture.
- Click to capture z-stack images for each worm.
NOTE: All following steps may be performed at any time.
3. Prepare Dopaminergic Neuron Images for Scoring
- For each z-stack, open the image file using either the microscope software or an external image analysis software, load the stack in the software, and compress the stack into a single flattened image.
- Blind images between and within treatment groups manually or using automatic blinding software.
4. Score Dopaminergic Neuron Dendrites
- Work with one neuron image at a time. Choose one of the four CEP dendrites to assess for blebs, breaks, and irregularities including bends, kinks, and curves. Scoring from the left to right when the nose is at the top of the image is recommended to ensure repeatability in scoring.
- Using the following guidelines, assign one score value to the dendrite. See Figure 1 for representative scoring image examples.
0- no damage, "perfect" neurons
1- irregular (kinks, curves, etc.)
2- <5 blebs
3- 5-10 blebs
4- >10 blebs and/or breakage removing <25% of total dendrite
5- breakage, 25-75% of dendrite removed
6- breakage, >75% dendrite removed- If multiple criteria are met within a single dendrite (i.e., kinks and blebs), assign the highest applicable score.
- Do not score dendrites that are not clearly visible, due to issues with image capture, overlapping with other dendrites, etc. If zooming in on a flattened z stack image, be mindful of enlarged pixels resembling false blebs.
- Repeat for each dendrite. Repeat for all images.
- Record all scores. Scores may be un-blinded at this time.
5. Prepare and Present Data
- Calculate the total number of dendrites in each treatment group assigned to each neurodegeneration score. Calculate the total number of scored dendrites in each treatment group.
- Divide the neurodegeneration score tallies by the total number of dendrites scored in the treatment group. Present data as a proportion of dendrites within a treatment group at each neurodegeneration score.
6. Perform Statistical Analysis
- Using a programming software or manually, run a chi-squared test for independence between all treatment group pairs to be compared. When appropriate, apply a Bonferroni correction of the p-value according to the number of compared experimental groups to account for multiple comparisons.
NOTE: This test will determine significant differences between two groups, but details of the type of difference must be qualified by eye.- Select comparisons between experimental groups. This will vary based on experimental design.
NOTE: In our experiments, typically, controls are compared to their respective treatment groups, all controls are compared, and all treatments are compared.
- Select comparisons between experimental groups. This will vary based on experimental design.
7. Practice Scoring with Practice Set of Dopaminergic Neuron Images
- See Supplementary File 1 for a set of neuron images presenting across the full range of our scoring system with commentary and score key. This practice set is intended to train researchers new to this method and ensure inter-rater reliability.
8. Consider Alternate Protocol Options
- Instead of capturing z-stacks, complete scoring at the microscope, without saving or stacking images.
NOTE: This option reduces requirements for technology capabilities, but removes the option for creating an archive of neuron images to return to at a later time, requires manual blinding, and permits blinding only between and not within treatment groups. - Instead of creating a single compressed image per stack, complete scoring by scrolling through the images of each z-stack.
NOTE: This option may be easier for some scorers and it reduces the risk for seeing false blebs on worms that moved during imaging and allows for scoring overlapping dendrites, but requires manual blinding and permits blinding only between and not within treatment groups.
Results
The scoring system described here was used to assess neurodegeneration in L4 larval stage BY200 [vtIs1 (dat-1p::GFP, rol-6)] C. elegans after rotenone exposure. Results of this experiment are shown in Figure 2 and represent our scoring system's ability to detect and quantify variable levels of dopaminergic neuron damage. Rotenone is a naturally occurring electron transport chain complex I inhibitor used in some pesticides, piscicides, and insecticides36,37. Note that working with toxic chemicals such as rotenone is inherently dangerous, and all labs should comply with all use and disposal regulations set by their institutions. In this experiment, liquid rotenone exposures at two doses, 0.03 µM and 0.5 µM, along with a control group, were begun immediately following a 0.5 M sodium hydroxide/1% sodium hypochlorite lysis to harvest eggs38. Eggs hatched in complete K-medium33,39 with 0.25% dimethyl sulfoxide (DMSO), and worms remained in liquid for ~48 hours until mid-L4 larval stage, at which point they were removed from chemical exposure, and prepared, imaged, and, using Figure 1 as a reference, scored according to the protocol steps above. For the higher dose of 0.5 µM rotenone, eggs were harvested 24 hours in advance to account for a rotenone-induced developmental delay and ensure all worms were stage synchronized at the time of imaging.
Figure 2 further demonstrates how our laboratory visualizes data collected using this scoring system. In this figure, a dose-dependent neurodegeneration response can be appreciated, and the specific breakdown of the score distribution is displayed clearly. These particular results showcase how neuronal damage can present in different ways. For example, the 0.03 µM rotenone-exposed group has a decreased proportion of healthy neurons with a score of 0, as compared to the control group, yet also has a decreased proportion of 5 scores. Detecting this detail about the score distributions between experimental groups highlights the sensitivity of our seven-point scoring system. This data was analyzed for statistical significance according to the protocol, using a chi-squared test for independence with a Bonferroni correction.
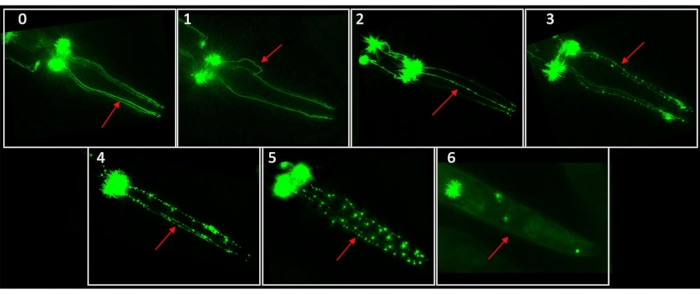
Figure 1. Dopamine neuron morphological alteration and degeneration scoring system representative images. This consolidated chart contains examples of neurons at each score and is intended to be used as a reference for scoring. Here, each labelled score corresponds to the most damaged dendrite in each worm, as indicated by the arrow in each panel. These images were taken using the protocol described in this paper with BY200 C. elegans. Please see Supplementary File 1 for a set of scored images with commentary to be used for training those new to this scoring method. Please click here to view a larger version of this figure.
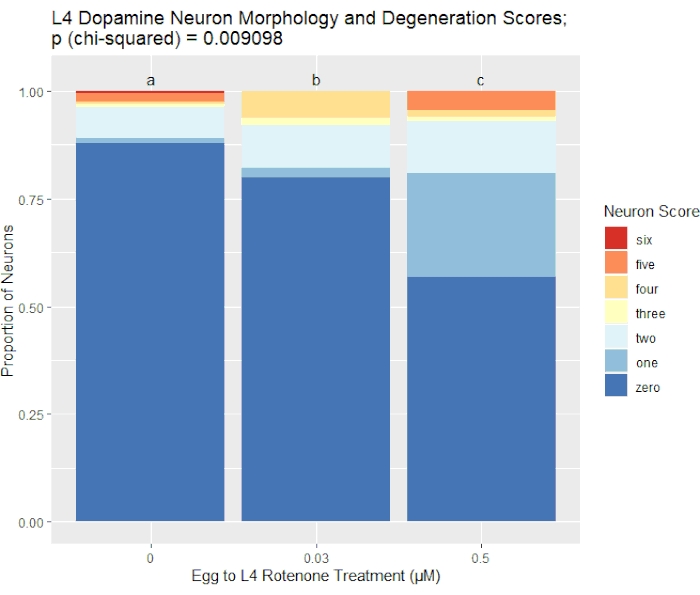
Figure 2. BY200 L4 dopamine neuron morphology and degeneration scores after rotenone exposure. This figure shows representative results analyzed using the scoring methods described here. The visualized greater proportions of damaged dopaminergic neurons with higher rotenone exposure concentrations were statistically analyzed using chi-squared tests for independence. Both rotenone treatment groups yielded statistically significant p-values when compared to the control group. Different letters indicate statistical difference. Please click here to view a larger version of this figure.
Supplementary File 1. Please click here to download this file.
Supplementary File 2. Please click here to download this file.
Discussion
This protocol demonstrates how to use the seven-point scale developed in our laboratory to quantify levels of dopaminergic neuron morphologic alteration and degeneration in C. elegans. We created and shared this scale as a tool to standardize analysis of dopaminergic neurodegeneration work in worms. Recognizing the importance of studying pathways involved in highly prevalent neurodegenerative diseases, many investigators take advantage of the C. elegans model's suitability for neurobiology visualization to study neurodegeneration29,31,32,33. However, there has yet to be an effort to reduce the large variation in how neuron damage is quantified across neurodegeneration research in worms. The scoring system presented here is thus intended to promote consistency in analyses and allow for comparison between studies.
Our scoring system may be used to analyze data derived from C. elegans experiments that use cell-specific fluorescent reporters that allow for visualization of dopaminergic neurons - specifically the CEP dendrites. Specifically, strains tagged at the dat-1 gene for GFP visualization of the dopaminergic neurons are compatible with this scoring protocol, though many other related transgenic models of PD do exist. It is possible that this scoring system would also be useful with those models; however, this should be validated prior to using them. In particular, it is possible (but not tested to our knowledge) strains with mCherry may not be well suited for this protocol as mCherry aggregation may be indistinguishable from blebs or lead to cell stress. Rather than providing a commentary on all specific models of PD and related neurodegenerative disorders, we focus on the scoring of neurodegeneration data itself. Additionally, this protocol focuses only on neuronal morphology and does not consider fluorescence levels of the soma. Neurodegeneration assays may be performed alongside behavioral assays relevant to neurodegenerative diseases, such as locomotion, lifespan, and health-span experiments. Levels of degeneration in established chemical, age-based, and genetic models of PD can also be confirmed and detailed using this scoring system. Measuring models, contributors, and pathways associated with PD and other neurodegenerative diseases can add to the scientific knowledge about these disorders and point towards how to manage the growing population of affected individuals. Having comparable neurodegeneration results across literature is key in supporting this goal.
To interpret the results derived from this scoring system, we propose considering each dendrite scored as n=1, because different neurons within the same worm often respond differently to treatment. This may be driven by the fact that only the CEPD neurons are directly exposed to the pseudocoelomic body fluid. As such, this allows the score spread of experimental groups to be displayed as proportions of the total number of dendrites scored in each group. This method, used for the representative results shown here, allows for easy comparison across treatment groups, accounts for differential responses within the same worm, and is easily analyzed with a Chi-squared test complimented by a Bonferroni correction for multiple comparisons. An example template for recording neuron scores and calculating percentages can be found in Supplementary File 2. We have considered two alternate methods for data analysis and identify flaws in each. The first option is averaging the scores of the four CEP neurons for each worm. This parametrizes the data; however, it assumes a linear relationship with increasing score and loses information about any variations in response to treatment within the same worm. The second option is to sum the scores of all four CEP neurons for each worm, which also parametrizes the data. This still assumes a linear relationship between scores, however it more capably accounts for differences within each worm than averaged scores by expanding the parameters of possible scores. However individual researchers decide to display their data, the results should be considered alongside experimental variables such as strain and worm age; for example, older worms have a higher expected baseline level of degeneration.
As these neurodegeneration score results are interpreted, researchers should also be aware of a few caveats and limitations of the scoring method. First, certain technological requirements are necessary to capture images suitable for scoring. The imaging microscope must support fluorescence channels and magnification and exposure settings that allow for clear visualization of CEP dendrites. As noted in the protocol, technological requirements may be reduced by protocol adjustments like scoring live images through the microscopic field rather than capturing images to be archived and scored at a later time. Second, possible statistical analysis methods for this data are limited as the data is non-parametric. The scoring scale is presumed to be progressive, but cannot be considered numeric since there are discrete score options and score increases are not necessarily proportional to each other with respect to biological function. For these reasons, chi-squared tests for independence are best suited for this type of data, meaning the statistical analysis depends on the observer to determine the direction of any statistical significance. Notably, the chi-squared test also only analyzes for differences in score distribution and is unable to provide evidence of differences in specific scoring categories. Finally, the functional significance of the morphological changes quantified by this scoring system have yet to be studied.
The future directions prompted by the development of this scoring system involve determining biological bases and correlations with individual neuron scores. Studying the functional significance (e.g. neuronal signaling, worm behavior) of all points on the scoring scale will inform how to better translate results to conclusions applicable to understanding the causes and consequences of neurodegenerative diseases and developing prevention and treatment options. Future research on neurodegeneration in worms should aim to discover connections to other morphology, such as worm shape and size. Additionally, neurodegeneration research can be supported by studying other reporter C. elegans strains to measure endpoints such as bioenergetics, reactive oxygen species production, and mitochondrial morphology.
Disclosures
The authors have no disclosures.
Acknowledgements
We wish to acknowledge Ian T. Ryde, for contributions to the development of the scoring scale and for his support during the creation of this manuscript. This work was supported by the National Institutes of Health (T32ES021432 supported KSM, and P42ES010356 to JNM).
Materials
| Name | Company | Catalog Number | Comments |
| 96-well plate | VWR | 29442-056 | For imaging in wells |
| Blinder | Solibyte Solutions LLC | Free software that blinds between and within uploaded sets of image files | |
| BY200 [vtIs1 (dat-1p::GFP, rol-6)] | Aschner Lab | C. elegans strain suitable for dopaminergic neuron fluorescent imaging. May be subsituted by other strains with a fluorescent reporter driven by cell-specific promotors Available upon request from the Meyer lab | |
| complete K-medium | 51 mM sodium chloride, 32 mM potassium chloride, 3 mM calcium chloride, 3 mM magnesium sulfate, 13 mM cholesterol | ||
| Coverslips 22x22mm, No.1 glass | VWR VistaVision | 48366-067 | For imaging on slides |
| dimethyl sulfoxide (DMSO) | Sigma-Aldrich | 472301 | Solvent for rotenone exposures |
| ImageJ | National Institutes of Health | ImageJ 1.5e or newer. Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, https:// imagej.nih.gov/ij/, 1997-2016. | Sotware for image manipulation |
| Keyence BZ-X All-in-one Fluoresence Microscope | Keyence | Used for fluorescent dopaminergic neuron image capture. May be substituted by other microscopes stuitable for fluorescent, high-resolution imaging | |
| Microscope Slides 3x1" | VWR VistaVision | 16004-420 | For imaging on slides |
| Rotenone | Sigma-Aldrich | R8875 | Electron transport chain complex I inhibitor |
| Sodium Azide (NaN_3) | Sigma-Aldrich | S2002 | Paralytic |
| Sodium Hydroxide | Sigma-Aldrich | S2770 | For bleach lysis |
| Sodium Hypochlorite | VWR | RC7495.5-32 | For bleach lysis |
| Tetramisole (Levamisole) Hydrochloride (HCl) | Sigma-Aldrich | L9756 | Paralytic |
References
- Maulik, M., Mitra, S., Bult-Ito, A., Taylor, B. E., Vayndorf, E. M. Behavioral Phenotyping and Pathological Indicators of Parkinson's Disease in C. elegans Models. Frontiers in Genetics. 8 (77), (2017).
- Hayes, M. T. Parkinson's Disease and Parkinsonism. Review. The American Journal of Medicine. 132 (7), 802-807 (2019).
- Pouchieu, C., et al. Pesticide use in agriculture and Parkinson's disease in the AGRICAN cohort study. International Journal of Epidemiology. 47 (1), 299-310 (2018).
- Brenner, S. The Genetics of Caenorhabditis elegans. Genetics. 77 (1), 71-94 (1973).
- Nass, R., Hall, D. H., Miller, D. M., Blakely, R. D. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 99 (5), 3264-3269 (2002).
- Wu, S., et al. Mutation of hop-1 and pink-1 attenuates vulnerability of neurotoxicity in C. elegans: the role of mitochondria-associated membrane proteins in Parkinsonism. Experimental Neurology. 309, 67-78 (2018).
- Chikka, M. R., Anbalagan, C., Dvorak, K., Dombeck, K., Prahlad, V. The Mitochondria-Regulated Immune Pathway Activated in the C. elegans Intestine Is Neuroprotective. Cell Reports. 16 (9), 2399-2414 (2016).
- Nass, R., Miller, D. M., Blakely, R. D. C-elegans: a novel pharmacogenetic model to study Parkinson's disease. Parkinsonism Relat D. 7 (3), 185-191 (2001).
- Benedetto, A., Au, C., Aschner, M., Nass, R., Nass, R., Przedborski, S. Manganese and C. elegans in Parkinson's disease. Parkinson's Disease: Pathogenic and Therapeutic Insights from Toxin and Genetic Models., Life Science. , (2008).
- Harrington, A. J., Hamamichi, S., Caldwell, G. A., Caldwell, K. A. C. elegans as a Model Organism to Investigate Molecular Pathways Involved with Parkinson's Disease. Developmental Dynamics. 239 (5), 1282-1295 (2010).
- Cooper, J. F., Van Raamsdonk, J. M. Modeling Parkinson's Disease in C. elegans. Journal of Parkinson's Disease. 8 (1), 17-32 (2018).
- Chege, P. M., McColl, G. Caenorhabditis elegans: a model to investigate oxidative stress and metal dyshomeostasis in Parkinson's disease. Frontiers in AGING NEUROSCIENCE. 6, 89 (2014).
- Lu, C. L., Svoboda, K. R., Lenz, K. A., Pattison, C., Ma, H. B. Toxicity interactions between manganese (Mn) and lead (Pb) or cadmium (Cd) in a model organism the nematode C. elegans. Environmental Science and Pollution Research. 25 (16), 15378-15389 (2018).
- Negga, R., et al. Exposure to Mn/Zn ethylene-bis-dithiocarbamate and glyphosate pesticides leads to neurodegeneration in Caenorhabditis elegans. Neurotoxicology. 32 (3), 331-341 (2011).
- Nagarajan, A., et al. Progressive degeneration of dopaminergic neurons through TRP channel-induced cell death. Journal of Neuroscience. 34 (17), 5738-5746 (2014).
- Salim, C., Rajini, P. S. Glucose-rich diet aggravates monocrotophos-induced dopaminergic neuronal dysfunction in Caenorhabditis elegans. Journal of Applied Toxicology. 37 (6), 772-780 (2017).
- Ved, R., et al. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. Journal of Biological Chemistry. 280 (52), 42655-42668 (2005).
- Yao, C., et al. LRRK2-mediated neurodegeneration and dysfunction of dopaminergic neurons in a Caenorhabditis elegans model of Parkinson's disease. Neurobiology of Disease. 40 (1), 73-81 (2010).
- Pivtoraiko, V. N., et al. Low-dose bafilomycin attenuates neuronal cell death associated with autophagy-lysosome pathway dysfunction. Journal of Neurochemistry. 114 (4), 1193-1204 (2010).
- Civelek, M., Mehrkens, J. F., Carstens, N. M., Fitzenberger, E., Wenzel, U. Inhibition of mitophagy decreases survival of Caenorhabditis elegans by increasing protein aggregation. Molecular and Cellular Biochemistry. 452 (1-2), 123-131 (2019).
- Van Pelt, K. M., Truttmann, M. C. Caenorhabditis elegans as a model system for studying aging-associated neurodegenerative diseases. Translational Medicine of Aging. 4, 60-72 (2020).
- Youssef, K., Tandon, A., Rezai, P. Studying Parkinson's disease using Caenorhabditis elegans models in microfluidic devices. Integrative Biology. 11 (5), 186-207 (2019).
- Lesage, S., Brice, A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Human Molecular Genetics. 18, 48-59 (2009).
- Gasser, T. Usefulness of Genetic Testing in PD and PD Trials: A Balanced Review. Journal of Parkinson's Disease. 5 (2), 209-215 (2015).
- Bronstein, J., et al. Meeting report: consensus statement-Parkinson's disease and the environment: collaborative on health and the environment and Parkinson's Action Network (CHE PAN) conference 26-28 June 2007. Environmental Health Perspectives. 117 (1), 117-121 (2009).
- Migliore, L., Coppede, F. Genetics, environmental factors and the emerging role of epigenetics in neurodegenerative diseases. Mutation Research. 667 (1-2), 82-97 (2009).
- Schapira, A. H. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurology. 7 (1), 97-109 (2008).
- Lill, C. M. Genetics of Parkinson's disease. Molecular and Cellular Probes. 30 (6), 386-396 (2016).
- Smith, L. Strengths and limitations of morphological and behavioral analyses in detecting dopaminergic deficiency in Caenorhabditis elegans. Neurotoxicology. 74, 209-220 (2019).
- Hindle, J. V. Ageing, neurodegeneration and Parkinson's disease. Age and Ageing. 39, 156-161 (2010).
- Luo, Z., et al. Age-dependent nigral dopaminergic neurodegeneration and α-synuclein accumulation in RGS6-deficient mice. JCI Insight. 4 (13), 126769 (2018).
- Berkowitz, L. A., et al. Video Article: Application of a C. elegans Dopamine Neuron Degeneration Assay for the Validation of Potential Parkinson's Disease Genes. Journal of Visualized Experiments. (17), e835 (2008).
- Tucci, M. L., Harrington, A. J., Caldwell, G. A., Caldwell, K. A. Modeling Dopamine Neuron Degeneration in Caenorhabditis elegans. Methods in Molecular Biology. 793 (19), 129-148 (2011).
- Hartman, J. H., et al. Genetic Defects in Mitochondrial Dynamics in Caenorhabditis elegans Impact Ultraviolet C Radiation- and 6-hydroxydopamine-Induced Neurodegeneration. International Journal of Molecular Sciences. 20 (3202), (2019).
- Caldwell, K. A., Wilicott, C. W., Caldwell, G. A. Modeling neurodegeneration in Caenorhabditis elegans. Disease Models & Mechanisms. 13, (2020).
- Cothren, S. D., Meyer, J. N., Hartman, J. H. Blinded Visual Scoring of Images Using the Freely-available Software Blinder. Biological Protocols. 8 (23), (2018).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 6758, Rotenone. PubChem. , (2021).
- Heinz, S., et al. Mechanistic Investigations of the Mitochondrial Complex I Inhibitor Rotenone in the Context of Pharmacological and Safety Evaluation. Scientific Reports. 7 (45465), (2017).
- Lewis, J. A., Fleming, J. T. Basic culture methods. Methods in Cell Biology. 48 (1), 3-29 (1995).
- Boyd, W. A., et al. Application of a Mathematical Model to Describe the Effects of Chlorpyrifos on Caenorhabditis elegans Development. PLoS ONE. 4 (9), 7024 (2009).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved