Method Article
Detection of Neutralization-sensitive Epitopes in Antigens Displayed on Virus-Like Particle (VLP)-Based Vaccines Using a Capture Assay
In This Article
Summary
Here, we present a protocol to detect neutralization epitopes on antigen-displaying virus-like particles (VLPs). Immunoprecipitation of the human immunodeficiency virus (HIV)-derived VLPs is performed using envelope glycoproteins-specific monoclonal antibodies coupled to protein G-conjugated magnetic beads. Captured VLPs are subsequently subjected to SDS-PAGE and Western blot-analysis employing viral core protein Gag-specific antibodies.
Abstract
The virus-like particle (VLP) capture assay is an immunoprecipitation method, commonly known as a 'pull-down assay' used to purify and isolate antigen-displaying VLPs. Surface antigen-specific antibodies are coupled to, and thus immobilized on a solid and insoluble matrix such as beads. Due to their high affinity to the target antigen, these antibodies can capture VLPs decorated with the cognate antigen anchored in the membrane envelope of the VLPs. This protocol describes the binding of antigen-specific antibodies to protein A- or G-conjugated magnetic beads. In our study, human immunodeficiency virus (HIV)-derived VLPs formed by the group-specific antigen (Gag) viral core precursor protein p55 Gag and displaying the envelope glycoproteins (Env) of HIV are examined. The VLPs are captured utilizing broadly neutralizing antibodies (bNAbs) directed against neutralization-sensitive epitopes in Env. The VLP capture assay outlined here represents a sensitive and easy-to-perform method to demonstrate that (i) the VLPs are decorated with the respective target antigen, (ii) the surface antigen retained its structural integrity as demonstrated by the epitope-specific binding of bNAbs used in the assay and (iii) the structural integrity of the VLPs revealed by the detection of Gag proteins in a subsequent Western blot-analysis. Consequently, the utilization of bNAbs for immunoprecipitation facilitates a prediction of whether VLP vaccines will be able to elicit a neutralizing B cell response in vaccinated humans. We anticipate that this protocol will furnish other researchers with a valuable and straightforward experimental approach to examine potential VLP-based vaccines.
Introduction
Virus-like particles (VLPs) resemble the native virus particle structure while lacking the viral genome, thus providing a high safety profile1,2. VLPs represent an individual class of vaccines increasingly developed due to their high immunogenicity3,4,5,6,7. This is particularly the case for membrane-enveloped VLPs, allowing for the display of not only homologous viral surface antigens but also heterologous antigens such as tumor antigens8,9,10. Figure 1 provides an exemplary overview of the structure of an enveloped antigen-decorated VLP. During the development process of VLP-based vaccines, assays are indispensable enabling the analysis of the respective target antigen displayed on the VLP surface. Such assays should be instrumental to elucidate the composition of a particulate vaccine: (i) Are the VLPs decorated with the respective surface antigen? (ii) Has the surface antigen retained its native structure as demonstrated by epitope recognition of neutralizing antibodies (bNAbs) and (iii) can the structural integrity of the VLPs be confirmed due to the detection of the viral protein mediating VLP formation?
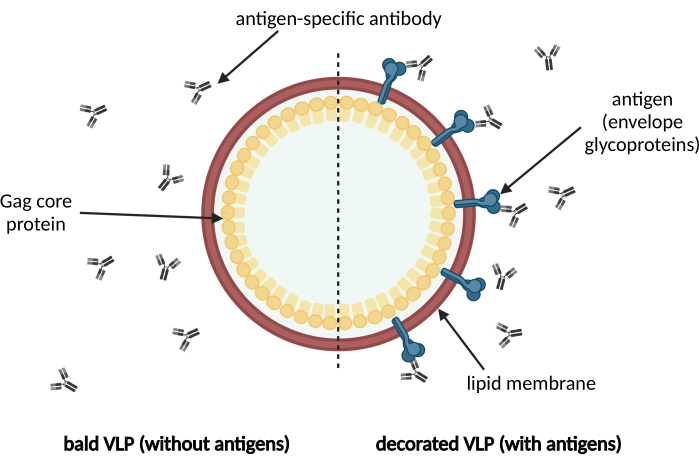
Figure 1: Schematic illustration of a membrane-enveloped VLP. VLPs are formed by immature precursor Gag core proteins and surrounded by a lipid membrane derived from the host cell. The antigens, e.g., envelope glycoproteins, are incorporated into the lipid membrane and displayed on the surface of the VLP (on the right). Antigen-specific antibodies recognize the antigen. On the left, a bald VLP without antigen decoration is shown. Please click here to view a larger version of this figure.
Especially VLPs formed by the viral group-specific antigen (Gag) core precursor protein p55 of human immunodeficiency virus type 1 (HIV-1) are preferred scaffolds for antigen display in vaccine development as numerous antibodies, and ELISA kits are available, enabling the quantification of these VLPs11,12. The HIV-1 envelope glycoproteins (Env), namely, the transmembrane protein gp41 (gp41-TM) and the soluble surface unit gp120 (gp120-SU) forming heterodimers, are incorporated into the membrane envelope of particles and are crucial target antigens for the development of vaccines against HIV infection13,14,15. The display of neutralization-sensitive epitopes in these target antigens is a prerequisite for eliciting a broadly neutralizing antibody response in vaccinees. Besides a T cell response directed against the Gag proteins, this is considered an important correlate of protection against HIV infection16. Consequently, and upon design and production of VLPs decorated with target antigen candidates, the subsequent analysis of the quality of the displayed antigens represents a critical step in the process of vaccine development.
Immunoprecipitation (IP) is a widely used technique for the detection of protein-protein interactions and the purification of protein complexes at a small scale17. Barret et al. first reported on the development of IP in 1960, yet, this method has been constantly further improved. IP enables the capture and isolation of a target antigen (prey) from a solution by employing an antigen-specific antibody (bait) immobilized by coupling to beads18,19. In this protocol, we demonstrate a variation of the classical IP application using membrane-enveloped p55 Gag-formed VLPs as prey and bNAbs that recognize neutralization-sensitive epitopes in the envelope proteins displayed on the surface of the VLPs as bait proteins. The successful application of this VLP capture assay facilitates the prediction of whether the tested antigen-positive VLPs will be able to elicit a neutralizing B cell response in vaccinated people. Such immunogenic properties of VLP-based vaccine candidates are frequently demonstrated in small animal models20,21,22.
In order to assess the quality of the newly developed VLP vaccine candidate, VLP capture assays have been successfully used5,23,24. However, the number of published methods is limited. The VLP capture assay presented here starts with the immobilization of Env-specific bNAbs on protein G-conjugated beads, which bind to the Fc region of mammalian-derived antibodies. Typical matrices for the immobilization of the antibody of choice are agarose or magnetic beads. However, magnetic beads are favorable for high-throughput applications25. In the next step, VLPs displaying the target antigen are captured by bNAb-coated beads. The formed immune complexes consisting of Env-positive VLPs and immobilized bNAbs are easily enriched using a magnet. The isolated immune complexes are eluted in the final step. Subsequently, the VLPs can be biochemically characterized. Here, we performed Western blot-analysis employing p55 Gag viral core protein-specific antibodies to demonstrate that the precipitated target Env antigens were not only harboring the neutralization-sensitive epitopes but were also displayed on Gag-formed VLPs. Furthermore, the detection of the viral core Gag proteins increases the sensitivity of the capture assay since Gag proteins are more abundant than Env in a VLP. In HIV-1, Env proteins are only present at a single- or double-digit number26, whereas more than 3,500 Gag molecules form the core of a particle27.
Compared to other techniques for the examination of protein-protein interactions28,29, the VLP capture assay provides an alternative method for research laboratories not having access to expensive analytical instruments. For example, transmission electron microscopic-analysis (TEM), surface plasmon resonance spectroscopy (SPR), and nanoparticle tracking-analysis (NTA) can be cost-intensive. The capture assay presented here also allows later subjection of captured antigen-positive VLP samples to further protein characterization, e.g., employing gel electrophoresis, immunoblotting, electron microscopy, and mass spectrometry (MS), respectively. Considering that the native structure of the target antigen is preserved during the VLP capture assay, also the performance of a native PAGE and subsequent immunoblotting techniques can be utilized.
The VLP capture assay represents an easy-to-use and sensitive method to examine the decoration of VLPs with target antigens exposing neutralization-sensitive epitopes, and thus their utility as future vaccine candidates.
Protocol
1. Sample preparation
- Seed the VLP producer suspension cell lines derived from 293-F cells expressing HIV structural genes gag alone or in concert with env 30 at a low cell density of 0.5 x 106 cells per mL in 293-F Expression Medium.
- Let them expand for 3-4 days at 37 °C and 8% CO2 in a shaker incubator rotation with an orbit of 5 cm and 135 rounds per minute (rpm).
- Pellet the producer cells by centrifugation at 100 x g for 5 min. Filter the clarified supernatant to remove residual cells and cell debris using 0.45 µm polyvinylidene fluoride (PVDF) membrane syringe filters to obtain cell-free cell culture supernatant (CFSN).
NOTE: The protocol can be paused here. CFSN can be stored overnight at 4 °C. - Either use the CFSN directly for the experiment or pellet VLPs from 35 mL of CFSN employing ultracentrifugation (112,700 x g, 4 °C, 1.5 h).
- Upon ultracentrifugation, discard the supernatant and suspend the VLP pellets in 200 µL of 15% (w/v) trehalose solution per centrifuge tube.
NOTE: The VLP pellet is often not visible to the naked eye. The protocol can be paused here. VLPs can be stored at -80 °C. - Prior to the VLP capture assay, determine the viral core protein concentrations in the VLP containing samples using an ELISA. This step is important to standardize the capture assay input.
2. VLP capture assay
NOTE: An overview of the workflow is depicted in Figure 2.
- Suspend the magnetic beads by either pipetting up and down or mixing on a rotator at 50 rpm for at least 5 min.
- Meanwhile, prepare the antibody solution containing the bNAbs. Use 10 µg of each bNAb in 200 µL of antibody binding and washing buffer (see Table of Materials) per reaction.
NOTE: Antibody amounts may vary depending on the affinity of the bNAb and antigen decoration density on the VLPs used. - Use 50 µL of the magnetic bead solution (see Table of Materials) per reaction and transfer the beads into a 1.5 mL reaction tube. Place the tubes on the magnetic separation rack.
NOTE: Alternatively, a strong single magnet can be used for each tube to separate the beads from the supernatant. - Wait a few minutes until the beads gather at the tube wall to ensure that all beads are collected. Remove the supernatant.
- Remove the magnet and suspend the beads in 200 µL of the bNAb solution prepared in step 2.2. Incubate for 30 min to 3 h mixing on a rotator at 50 rpm at room temperature.
- Place the reaction tubes in the magnetic separation rack again, wait and remove the supernatant.
- Remove the tubes from the magnet and wash the beads by resuspending in 200 µL of antibody binding and washing buffer.
- Repeat step 2.4. Remove as much washing buffer as possible.
- Add the samples to the bead-bound bNAbs.
NOTE: The VLP input for the capture assay using HIV-derived particles should be at least 15 ng of viral core protein per reaction. VLP amounts may vary depending on the affinity of the bNAb, and the antigen decoration density on the VLPs used. - If the sample volume added in step 2.9 is below 1 mL, add PBS to adjust the sample volume to 1 mL. Suspend the beads gently by pipetting.
- Incubate the samples and beads for 2.5 h on a rotator at room temperature. Ensure that the beads stay in suspension and the solution is thoroughly mixed during incubation.
- Place the tubes on the magnet and remove the supernatant.
- Wash the magnetic beads by suspending them in 200 µL of washing buffer (see Table of Materials). Repeat the washing step three times.
- Suspend the beads in 100 µL of washing buffer and transfer the suspension to a clean (heat resistant) reaction tube.
- Place the tube on the magnetic separation rack (see Table of Materials) and remove the supernatant completely.
- Prepare denatured SDS-PAGE samples following steps 2.16.1-2.16.2 or non-denatured samples by eluting the VLPs from the magnetic beads following steps 2.16.3-2.16.5.
- To prepare denatured SDS-PAGE samples, suspend the beads in 20-80 µL of Laemmli buffer (0.8 µL of Laemmli buffer per 1 ng of p55 Gag assay input) and incubate at 95 °C for 5 min.
- Proceed directly with SDS-PAGE or store the samples at -20 °C.
CAUTION: Laemmli buffer contains 2-mercaptoethanol and sodium dodecyl sulfate. Wear protective gloves and eye protection. Avoid contact with skin, eyes, and clothes. Do not inhale vapors. Work in a well-ventilated space, e.g., a fume hood.
NOTE: The protocol can be paused here. - Alternatively, perform a non-denaturing elution step to obtain VLPs with preserved native protein structure.
- Add 25 µL of elution buffer (frequently provided with the beads) to the magnetic beads and incubate for 2-5 min at room temperature.
- Remove the beads and transfer the eluate to a clean tube. Use the eluate for subsequent assays or store it at 4 °C.
NOTE: The protocol can be paused here.
3. Analysis of captured VLP samples
NOTE: For analysis of the captured VLPs, conduct an SDS-PAGE and subsequently Western blot-analysis31,32.
- Place the sample tubes on the magnetic rack to separate the beads from the solution.
- Load 10 µL of each sample into individual wells of the gel.
NOTE: In this protocol, viral p55 Gag core proteins were detected by employing polyclonal rabbit antibodies directed against HIV Gag and secondary polyclonal chicken anti-rabbit IgG coupled to horseradish peroxidase (HRP) antibodies. Viral core proteins were visualized using chemiluminescence detection.
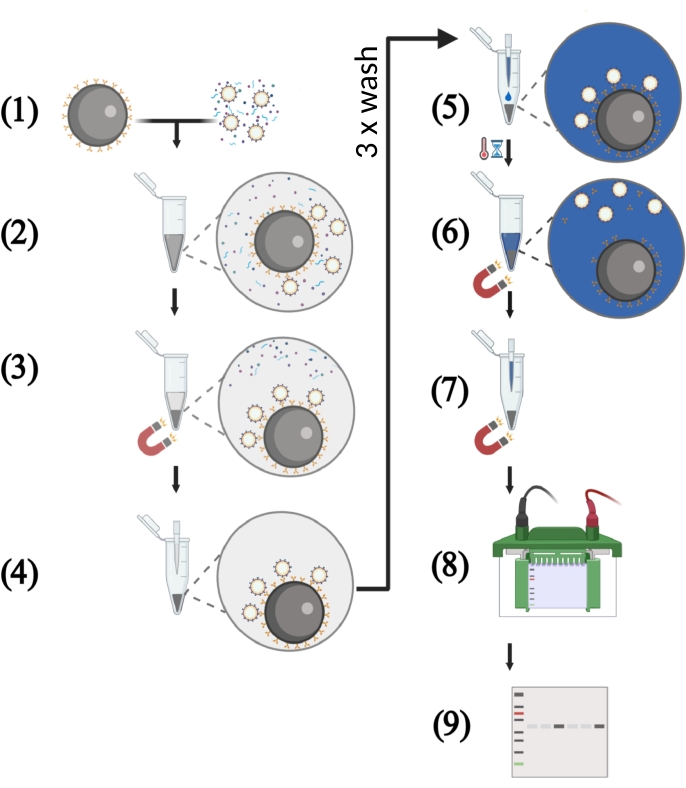
Figure 2: Schematic illustration of key steps of the VLP capture assay using cell-free supernatants. (1) The VLP capture assay starts with the binding of the HIV-1 bNAbs to the protein G-coupled magnetic beads. Meanwhile, the VLP solution with a defined p55 concentration is prepared. The cell-free culture supernatant consists of a mixture of p55 Gag VLPs, host cell proteins, and nucleic acids. (2) The magnetic beads coupled to bNAbs and the VLP solution are transferred into a 1.5 mL reaction tube followed by incubation under rotation. (3) Antigen-displaying VLPs are captured by the bNAbs-coated beads. Separation of these immune complexes from host cell-derived contaminants is performed in a magnetic field. (4) The supernatant is removed by pipetting, and the beads are washed three times to remove unbound VLPs. (5) In the next step, reducing protein loading buffer is added to the immune complexes consisting of magnetic beads coated with bNAbs and captured VLPs. (6) Boiling the sample dissociates bNAbs and target antigen from the magnetic beads and lysis the VLPs. (7) Magnetic beads are separated from the solution in a magnetic field. (8) The protein samples are subjected to SDS-PAGE. (9) Western blot-analysis is performed in order to detect viral p55 Gag core proteins. Please click here to view a larger version of this figure.
Results
Figure 3 shows a representative outcome of VLPs first captured from CFSN and VLP pellets, respectively, using bNAbs and subsequently subjected to Western blot-analysis to detect viral core proteins. The beads used for the capture assay were coated with three different bNAbs directed against neutralization-sensitive epitopes of the Env glycoproteins and isotype antibodies serving as negative controls, respectively. Using isotype antibodies-coated beads, no Gag proteins were detectable in samples containing Env-negative (bald VLPs) or Env-displaying VLPs employing Western blot-analysis. This demonstrated that the unspecific binding of VLPs to beads coated with human antibodies did not mediate VLP capture. Bald VLPs were also not bound by bNAbs-coated beads, and consequently, again, no Gag proteins were detectable. In contrast, all three bNAbs captured VLPs displaying Env proteins (Env VLPs), and therefore, Gag proteins were subsequently readily detected. As visible in Figure 3, the capture assay can be utilized to analyze target antigen displaying VLPs in CFSN and pelleted VLP samples, respectively.
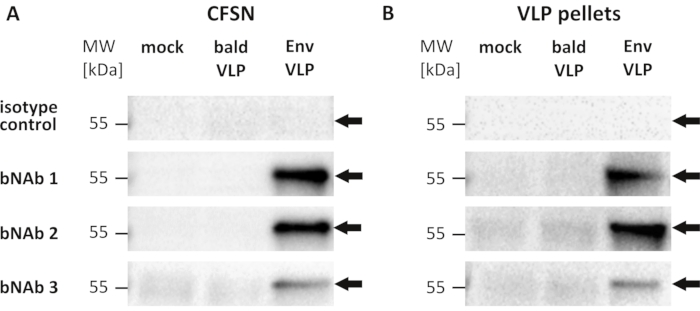
Figure 3: Detection of neutralization-sensitive epitopes in HIV-1 envelope glycoproteins displayed on p55 Gag-formed VLPs using broadly neutralizing antibodies. Representative results of Western blot-analysis using viral Gag core protein-specific antibodies after performing a VLP capture assay employing three different broadly neutralizing antibodies (bNAb 1, bNAb 2, and bnAb 3) targeting epitopes within the HIV-1 envelope glycoproteins (Env). Isotype human antibodies pooled from human sera served as negative controls. Cell culture supernatants were harvested from suspension cell cultures producing VLPs with Env proteins (Env VLPs) and bald VLPs (Env-negative), respectively, as well as from naive cells not expressing any viral proteins (mock). Both the supernatants from the bald VLP producer cell culture as well as from the mock cell culture served as negative controls. Supernatants were freed from contaminating cells using low-speed centrifugation and subsequent filtration to obtain cell-free supernatants (CFSN) for analysis. Upon immunoprecipitation, Western blot-analysis was performed using polyclonal rabbit antibodies directed against Gag and secondary anti-rabbit IgG-HRP conjugates. The apparent molecular weight in kilodaltons (kDa) as visible from the molecular weight marker (MW) is depicted on the left. The arrows indicate the detected precursor protein p55 Gag. (A) For each sample, CFSN containing 100 ng of Gag protein was applied to the capture assay to standardize the input amount of VLPs. CFSN was directly incubated with antibody-coated beads. (B) VLP pellets were obtained by ultracentrifugation of CFSN. Pellets containing 100 ng of Gag protein were applied to the capture assay using isotype antibodies and bNAb 3. Samples for bNAb 1 and bNAb 2 contained only 25 ng of Gag. Please click here to view a larger version of this figure.
Discussion
Prior to the VLP capture assay, evaluate the formation of VLPs and the expression of the target antigen in the VLP producer cell lines. Instrumental methods are flow cytometric-analysis of the cell surface expression of the antigen as well as antigen- and viral core protein-specific ELISA of CFSN and pelleted VLPs.
Critical steps of the VLP capture assay are the coating of the beads with capture antibodies - here bNAbs - and the subsequent capture of the antigen-positive VLPs by the antibody-coated beads. Successful coating of the beads with antibodies depends on the choice of the conjugated immunoglobulin (Ig)-binding protein. The donor species as well as the Ig class of the antibodies determine whether protein G- or protein A-conjugated beads are preferable. For most species and Ig classes, protein G is the ligand of choice33. As an alternative to protein A/G-conjugated beads, streptavidin beads for the coating with biotinylated antibodies are available. Beads can also be covalently coupled with antibodies.
The capture of the VLPs by antibody-coated beads depends on thorough mixing, sufficient incubation time, antigen abundance, and affinity of the capture antibody. In our experience, thorough mixing of the antibody-coated beads with the VLP samples is best achieved by the utilization of volumes >500 µL in 1.5 mL tubes under rotation for at least 2 h at room temperature or 4 °C. Another potential hurdle is the too low amount of VLPs in the sample. For antibodies strongly binding the target antigen, VLP inputs as low as 15 ng of Gag protein usually allow for readily detectable amounts of the viral core proteins utilizing Western blot-analysis. However, low-affinity antibodies require higher input amounts, e.g., 100 ng of Gag protein, to obtain conclusive results (Figure 3, bNAb 3).
Some surface antigens are prone to protease degradation. Here, we recommend the addition of protease inhibitors to the VLP samples and incubation at 4 °C. Non-specific adhesion of host cell proteins and VLPs to the bead-bound antibodies is rarely observed and should be excluded by utilizing appropriate negative controls, as we demonstrated here using mock and bald VLP samples and isotype control antibodies. Strategies to reduce non-specific binding include extended washing steps and the addition of casein in the washing buffer34. Furthermore, the capture assay may also be improved by determining the optimal ratio of antibody to antigen-displaying VLP amount.
In the last step of the VLP capture assay, we describe the elution of the immune complexes from the beads by boiling in reducing Laemmli buffer. During this step, the VLPs are disassembled, and the capture antibodies and target antigens are separated from the beads. Notably, the donor species of the primary antibody used in the subsequent Western blot-analysis has to differ from the donor of the capture antibody to avoid the unintended detection of the capture antibody by the secondary anti-donor IgG HRP-conjugated antibodies.
The VLP capture assay presented here provides an easy-to-use and sensitive method to detect neutralization-sensitive epitopes in structural intact target antigens displayed on VLP surfaces. However, the capture assay does not enable direct epitope quantification. ELISA performed with bNAbs are instrumental for this purpose and should be conducted in parallel, particularly if examined VLPs are intended to be used in preclinical studies employing animal models35. This is pivotal, as the amount of antigen can directly correlate with the elicitation of a neutralizing antibody response in immunized animals, as shown for porcine circovirus type 2 (PCV2) vaccines36.
An ideal vaccine should result in the elicitation of bNAbs targeting the neutralization-sensitive epitopes on the virion surface. The analysis of these epitopes especially referring to their full structural integrity on the particulate vaccine surface is crucial for identifying potential vaccine candidates. This is not only the case for HIV-derived VLPs but also for many other VLP vaccines in development37. Prominent VLP-based vaccines are, for example, derived from non-enveloped or capsid parental viruses such as human papillomavirus (HPV). Unlike HIV-1 particles, which are formed by only one structural core protein, namely p55 Gag, and enveloped by the membrane originating from the VLP producer cell, HPV particles consist of only one or two structural core proteins38,39. Likewise and as presented here for enveloped VLPs, the VLP capture assay may also be applicable to the detection of neutralization-sensitive epitopes of non-enveloped VLPs.
As an alternative to the capture assay, VLP samples can be directly subjected to native PAGE followed by Western blot-analysis using bNAbs and appropriate secondary antibodies coupled to HRP40. However and for the analysis of HIV Env-decorated VLPs, this assay is less sensitive as only a low number of antigen proteins per VLP can be expected. In contrast, the capture assay facilitates the detection of the core proteins abundant at large amounts per VLP, in the case of HIV-derived VLPs more than 3,500 Gag proteins form a VLP27. This allows for the very sensitive indirect detection of epitopes in Env displayed even at low densities on VLPs.
The number of well-established methods to examine neutralization-sensitive epitopes in surface antigens of VLPs is limited. Labeling the antigens displayed on the VLPs is possible with epitope-specific antibody-fluorophore conjugates and subsequent detection by nanoparticle tracking-analysis (NTA), enabling detection and quantification of VLPs. This method has also been successfully developed and optimized for exosomes presenting cell surface markers41. Also, surface plasmon resonance (SPR) spectroscopy allows for the analysis of interactions between unconjugated neutralizing antibodies and cognate epitopes presented on VLPs. Although not suitable for higher throughput analysis, VLPs can also be labeled with bNAbs coupled to gold particles and subsequent transmission electron microscopic (TEM)-examination42.
In conclusion, the VLP capture assay provides some considerable advantages: (i) Assessment of the structural integrity of neutralization-sensitive epitopes on the surface of VLPs, (ii) sensitive and indirect detection of antigens even when displayed at low densities on VLPs, and (iii) the method does not require cost-intensive analytical equipment.
Disclosures
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by a grant from the German Federal Ministry of Education and Research, funding program Forschung an Fachhochschulen, contract numbers 13FH767IA6 and 13FH242PX6 to JS. Figures 1 and 2 were created with BioRender.com.
Materials
| Name | Company | Catalog Number | Comments |
| 1.5 mL reaction tubes | Eppendorf | ||
| 10x PBS | gibco | 70011044 | |
| 4%–15% Mini-PROTEAN TGX stain-free protein gels | BioRad | 4568085 | |
| Antibodies (bnAbs) | Polymun Scientic | ||
| Isotype control antibody | invitrogen (Thermo Fisher Scientific) | 02-7102 | |
| Chemidoc XRS+ imaging system | BioRad | 1708265 | |
| Chicken anti-rabbit IgG HRP-coupled | Life technologies | A15987 | 1:5000 in TBS-T + 2 % (w/v) powdered milk |
| Dynabeads Protein G Immunoprecipitation Kit | invitrogen (Thermo Fisher Scientific) | 10007D | includes buffers and washing solutions |
| FreeStyle 293-F cells | invitrogen (Thermo Fisher Scientific) | R790-07 | |
| FreeStyle 293 Expression Medium | invitrogen (Thermo Fisher Scientific) | 12338026 | |
| Gel blotting papers | Whatman | GB005 | |
| Glycine | Carl Roth | 0079 | blotting buffer |
| Magnetic separation rack | New England Biolabs | S1509S | for 12 x 1.5 mL or 6 x 1.5 mL tubes |
| Methanol | Carl Roth | 4627 | blotting buffer |
| Mini-PROTEAN Tetra Cell electrophoresis system | BioRad | ||
| Optima XE-90 ultracentrifuge | Beckman Coulter | ||
| PageRuler prestained protein ladder | Thermo Scientific | 26616 | |
| Polyvinylidene fluoride (PVDF) syringe filters, 0.45 µm | Carl Roth | KC89.1 | |
| Powdered milk | Carl Roth | T145 | blocking buffer |
| PVDF transfermembrane, 0.45 µm | Carl Roth | T830.1 | |
| QuickTiter HIV p24 ELISA | Cell Biolabs | VPK-108-H | |
| Rabbit polyclonal to HIV1 p55 + p24 + p17 | abcam | ab63917 | 1:2000 in TBS-T + 2 % (w/v) powdered milk |
| Rotator | Heidolph | REAX2 | |
| ROTI Load 1 (laemmli buffer) | Carl Roth | K929.1 | 4x concentrated reducing protein gel loading buffer |
| ROTIPHORESE 10x SDS-PAGE | Carl Roth | 3060 | |
| Sodium chloride | Carl Roth | 3957 | TBS-T buffer |
| SuperSignal West Pico PLUS chemiluminescent substrate | Thermo Scientific | 34579 | |
| SW28 rotor | Beckman Coulter | ||
| Thermomixer | Cel Media | basic | |
| Trans-Blot Turbo | BioRad | ||
| Trehalose dihydrate | Carl Roth | 8897.2 | |
| TRIS | Carl Roth | 5429 | blotting buffer |
| TRIS hydrochloride | Carl Roth | 9090 | TBS-T buffer |
| Tween-20 | Carl Roth | 9127 | TBS-T buffer |
| Ultra Clear centrifuge tubes | Beckman Coulter | 344058 |
References
- Roldão, A., Mellado, M. C. M., Castilho, L. R., Carrondo, M. J. T., Alves, P. M. Virus-like particles in vaccine development. Expert Review of Vaccines. 9 (10), 1149-1176 (2010).
- Noad, R., Roy, P. Virus-like particles as immunogens. Trends in Microbiology. 11 (9), 438-444 (2003).
- Qian, C., et al. Recent progress on the versatility of virus-like particles. Vaccines. 8 (1), 139 (2020).
- Zabel, F., Kündig, T. M., Bachmann, M. F. Virus-induced humoral immunity: On how B cell responses are initiated. Current Opinion in Virology. 3 (3), 357-362 (2013).
- Garg, H., Mehmetoglu-Gurbuz, T., Joshi, A. Virus Like Particles (VLP) as multivalent vaccine candidate against Chikungunya, Japanese Encephalitis, Yellow Fever and Zika Virus. Scientific Reports. 10 (1), 4017 (2020).
- Hodgins, B., Pillet, S., Landry, N., Ward, B. J. A plant-derived VLP influenza vaccine elicits a balanced immune response even in very old mice with co-morbidities. PLoS ONE. 14 (1), 0210009 (2019).
- Lai, C. C., et al. Process development for pandemic influenza VLP vaccine production using a baculovirus expression system. Journal of Biological Engineering. 13, 78 (2019).
- Caldeira, J. C., Perrine, M., Pericle, F., Cavallo, F. Virus-like particles as an immunogenic platform for cancer vaccines. Viruses. 12 (5), 488 (2020).
- Nika, L., et al. An HER2-displaying virus-like particle vaccine protects from challenge with mammary carcinoma cells in a mouse model. Vaccines. 7 (2), 41 (2019).
- Mohsen, M. O., Zha, L., Cabral-Miranda, G., Bachmann, M. F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Seminars in Immunology. 34, 123-132 (2017).
- Fontana, D., Garay, E., Cevera, L., Kratje, R., Prieto, C., Gòdia, F. Chimeric VLPs based on hiv-1 gag and a fusion rabies glycoprotein induce specific antibodies against rabies and foot-and-mouth disease virus. Vaccines. 9 (3), 251 (2021).
- Cervera, L., et al. Production of HIV-1-based virus-like particles for vaccination: achievements and limits. Applied Microbiology and Biotechnology. 103 (18), 7367-7384 (2019).
- Gonelli, C. A., King, H. A. D., Mackenzie, C., Sonza, S., Center, R. J., Purcell, D. F. J. Immunogenicity of HIV-1-based virus-like particles with increased incorporation and stability of membrane-bound env. Vaccines. 9 (3), 1-36 (2021).
- Trkola, A. HIV not as simple as one, two, three. Nature. 568, 321-322 (2019).
- Berman, P. W., et al. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature. 345 (6276), 622-625 (1990).
- Barouch, D. H. Challenges in the development of an HIV-1 vaccine. Nature. 455 (7213), 613-619 (2008).
- DeCaprio, J., Kohl, T. O. Immunoprecipitation. Cold Spring Harbor Protocols. 2020 (11), 449-461 (2020).
- Barret, B., Wood, P. A., Volwiler, W. Quantitation of gamma globulins in human serum by immunoprecipitation. Journal of Laboratory and Clinical Medicine. 55, 605-615 (1960).
- Kaboord, B., Perr, M. Isolation of proteins and protein complexes by immunoprecipitation. Methods in Molecular Biology. 424, 349-364 (2008).
- Lee, S. H., Chu, K. B., Kang, H. J., Quan, F. S. Virus-like particles containing multiple antigenic proteins of Toxoplasma gondii induce memory T cell and B cell responses. PLoS ONE. 14 (8), 0220865 (2019).
- Lee, Y. T., et al. Intranasal vaccination with M2e5x virus-like particles induces humoral and cellular immune responses conferring cross-protection against heterosubtypic influenza viruses. PLoS ONE. 13 (1), 0190868 (2018).
- Wang, J., et al. Large-scale manufacture of VP2 VLP vaccine against porcine parvovirus in Escherichia coli with high-density fermentation. Applied Microbiology and Biotechnology. 104 (9), 3847-3857 (2020).
- Swenson, D. L., et al. Generation of Marburg virus-like particles by co-expression of glycoprotein and matrix protein. FEMS Immunology and Medical Microbiology. 40 (1), 27-31 (2004).
- Latham, T., Galarza, J. M. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. Journal of Virology. 75 (13), 6154-6165 (2001).
- Doyle, J., Ray, M., Ouyang, A., Benton, B., Bell, P. A. Abstract 4877: High throughput proteomic applications using protein A/G magnetic beads. Association for Cancer Research (AACR) 102nd Annual Meeting. 20, 4877 (2011).
- Zhu, P., et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proceedings of the National Academy of Sciences of the United States of America. 100 (26), 15812-15817 (2003).
- Lavado-García, J., Jorge, I., Boix-Besora, A., Vázquez, J., Gòdia, F., Cervera, L. Characterization of HIV-1 virus-like particles and determination of Gag stoichiometry for different production platforms. Biotechnology and Bioengineering. 118 (7), 2660-2675 (2021).
- Miura, K. An overview of current methods to confirm protein-protein interactions. Protein & Peptide Letters. 25 (8), 728-733 (2018).
- Rao, V. S., Srinivas, K., Sujini, G. N., Kumar, G. N. Advances in molecular techniques to study diversity. Plant Biotechnology, Volume 1: Principles, Techniques, and Applications. , 341-365 (2017).
- Rosengarten, J. F., Schatz, S., Wolf, T., Barbe, S., Stitz, J. Components of a HIV-1 vaccine mediate virus-like particle (VLP)-formation and display of envelope proteins exposing broadly neutralizing epitopes. Virology. , 41-48 (2022).
- JoVE, Grundlegende Methoden in der Zell- und Molekularbiologie. Separating Protein with SDS-PAGE. JoVE Science Education Database. , (2021).
- Eslami, A., Lujan, J. Western blotting: sample preparation to detection. Journal of Visualized Experiments: JoVE. (44), e2359 (2010).
- Sheng, S., Kong, F. Separation of antigens and antibodies by immunoaffinity chromatography. Pharmaceutical Biology. 50 (8), 1038-1044 (2012).
- Guzzo, C., et al. Virion incorporation of integrin 47 facilitates HIV-1 infection and intestinal homing. Science Immunology. 2 (11), (2017).
- Wei, M., et al. Bacteria expressed hepatitis E virus capsid proteins maintain virion-like epitopes. Vaccine. 32 (24), 2859-2865 (2014).
- Jin, J., Park, C., Cho, S. H., Chung, J. The level of decoy epitope in PCV2 vaccine affects the neutralizing activity of sera in the immunized animals. Biochemical and Biophysical Research Communications. 496 (3), 846-851 (2018).
- Zhang, X., et al. Lessons learned from successful human vaccines: Delineating key epitopes by dissecting the capsid proteins. Human Vaccines and Immunotherapeutics. 11 (5), 1277-1292 (2015).
- DiGiuseppe, S., Bienkowska-Haba, M., Guion, L. G. M., Keiffer, T. R., Sapp, M. Human papillomavirus major capsid protein L1 remains associated with the incoming viral genome throughout the entry process. Journal of Virology. 91 (16), 00537 (2017).
- Wang, J. W., Roden, R. B. S. L2, the minor capsid protein of papillomavirus. Virology. 445 (1-2), 175-186 (2013).
- Binley, J. M., et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. Journal of Virology. 82 (23), 11651-11668 (2008).
- Thane, K. E., Davis, A. M., Hoffman, A. M. Improved methods for fluorescent labeling and detection of single extracellular vesicles using nanoparticle tracking analysis. Scientific Reports. 9 (1), 12295 (2019).
- Mulder, A. M., et al. Toolbox for non-intrusive structural and functional analysis of recombinant VLP based vaccines: A case study with hepatitis B vaccine. PLoS ONE. 7 (4), 0033235 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved