Method Article
Analyzing Mitochondrial Morphology Through Simulation Supervised Learning
In This Article
Summary
This article explains how to use simulation-supervised machine learning for analyzing mitochondria morphology in fluorescence microscopy images of fixed cells.
Abstract
The quantitative analysis of subcellular organelles such as mitochondria in cell fluorescence microscopy images is a demanding task because of the inherent challenges in the segmentation of these small and morphologically diverse structures. In this article, we demonstrate the use of a machine learning-aided segmentation and analysis pipeline for the quantification of mitochondrial morphology in fluorescence microscopy images of fixed cells. The deep learning-based segmentation tool is trained on simulated images and eliminates the requirement for ground truth annotations for supervised deep learning. We demonstrate the utility of this tool on fluorescence microscopy images of fixed cardiomyoblasts with a stable expression of fluorescent mitochondria markers and employ specific cell culture conditions to induce changes in the mitochondrial morphology.
Introduction
In this paper, we demonstrate the utility of a physics-based machine learning tool for subcellular segmentation1 in fluorescence microscopy images of fixed cardiomyoblasts expressing fluorescent mitochondria markers.
Mitochondria are the main energy-producing organelles in mammalian cells. Specifically, mitochondria are highly dynamic organelles and are often found in networks that are constantly changing in length and branching. The shape of mitochondria impacts their function, and cells can quickly change their mitochondrial morphology to adapt to a change in the environment2. To understand this phenomenon, the morphological classification of mitochondria as dots, rods, or networks is highly informative3.
The segmentation of mitochondria is crucial for the analysis of mitochondrial morphology in cells. Current methods to segment and analyze fluorescence microscopy images of mitochondria rely on manual segmentation or conventional image processing approaches. Thresholding-based approaches such as Otsu4 are less accurate due to the high noise levels in microscopy images. Typically, images for the morphological analysis of mitochondria feature a large number of mitochondria, making manual segmentation tedious. Mathematical approaches like MorphoLibJ5 and semi-supervised machine learning approaches like Weka6 are highly demanding and require expert knowledge. A review of the image analysis techniques for mitochondria7 showed that deep learning-based techniques may be useful for the task. Indeed, image segmentation in everyday life images for applications such as self-driving has been revolutionized with the use of deep learning-based models.
Deep learning is a subset of machine learning that provides algorithms that learn from large amounts of data. Supervised deep learning algorithms learn relationships from large sets of images that are annotated with their ground truth (GT) labels. The challenges in using supervised deep learning for segmenting mitochondria in fluorescence microscopy images are two-fold. Firstly, supervised deep learning requires a large dataset of training images, and, in the case of fluorescence microscopy, providing this large dataset would be an extensive task compared to when using more easily available traditional camera-based images. Secondly, fluorescence microscopy images require GT annotations of the objects of interest in the training images, which is a tedious task that requires expert knowledge. This task can easily take hours or days of the expert's time for a single image of cells with fluorescently labeled subcellular structures. Moreover, variations between annotators pose a problem. To remove the need for manual annotation, and to be able to leverage the superior performance of deep learning techniques, a deep learning-based segmentation model was used here that was trained on simulated images. Physics-based simulators provide a way to mimic and control the process of image formation in a microscope, allowing the creation of images of known shapes. Utilizing a physics-based simulator, a large dataset of simulated fluorescence microscopy images of mitochondria was created for this purpose.
The simulation starts with the geometry generation using parametric curves for shape generation. Emitters are randomly placed on the surface of the shape in a uniformly distributed manner such that the density matches the experimental values. A 3D point spread function (PSF) of the microscope is computed using a computationally efficient approximation8 of the Gibson-Lanni model9. To closely match the simulated images with experimental images, both the dark current and the shot noise are emulated to achieve photo realism. The physical GT is generated in the form of a binary map. The code for generating the dataset and training the simulation model is available10, and the step for creating this simulated dataset is outlined in Figure 1.
We showcase the utility of deep learning-based segmentation trained entirely on a simulated dataset by analyzing confocal microscopy images of fixed cardiomyoblasts. These cardiomyoblasts expressed a fluorescent marker in the mitochondrial outer membrane, allowing for the visualization of the mitochondria in the fluorescence microscopy images. Prior to conducting the experiment given as an example here, the cells were deprived of glucose and adapted to galactose for 7 days in culture. Replacing glucose in the growth media with galactose forces the cells in culture to become more oxidative and, thus, dependent upon their mitochondria for energy production11,12. Furthermore, this renders the cells more sensitive to mitochondrial damage. Mitochondrial morphology changes can be experimentally induced by adding a mitochondrial uncoupling agent such as carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to the cell culture medium13. CCCP leads to a loss of mitochondrial membrane potential (ΔΨm) and, thus, results in changes in the mitochondria from a more tubular (rod-like) to a more globular (dot-like) morphology14. In addition, mitochondria tend to swell during CCCP treatment15. We display the morphological distribution of mitochondria changes when the galactose-adapted cardiomyoblasts were treated with the mitochondrial uncoupler CCCP. The deep learning segmentations of the mitochondria enabled us to classify them as dots, rods, or networks. We then derived quantitative metrics to assess the branch lengths and abundance of the different mitochondrial phenotypes. The steps of the analysis are outlined in Figure 2, and the details of the cell culture, imaging, dataset creation for deep learning-based segmentation, as well as the quantitative analysis of the mitochondria, are given below.
Protocol
NOTE: Sections 1-4 can be omitted if working with existing microscopy images of mitochondria with known experimental conditions.
1. Cell culture
- Grow the H9c2 cells in DMEM without glucose supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM galactose, 10% FBS, 1% streptomycin/penicillin, and 1 µg/mL puromycin. Allow the cells to adapt to galactose for at least 7 days in culture before the experiments.
NOTE: The H9c2 cells used here have been genetically modified to express fluorescent mitochondria and have also acquired a puromycin resistance gene. The addition of this antibiotic ensures the growth of cells with the resistance gene and, thus, fluorescent mitochondria. - Seed the H9c2 cells for the experiment when the cell confluency reaches approximately 80% (T75 culture flask, assessed by brightfield microscopy). Pre-heat the medium and trypsin to 37°C for at least 15 min before proceeding.
NOTE: It is possible to perform the experiment in parallel with two or more different cell culture conditions. The H9c2 cell confluency should not be above 80% to prevent the loss of myoblastic cells. At 100% confluency, the cells form myotubes and start differentiating. - Prepare for seeding the cells, operating in a sterile laminar flow hood, by placing a #1.5 glass coverslip for each experimental condition in a well of a 12-well plate. Label each condition and the experimental details on the 12-well plate.
- Move the cell culture flask from the incubator to the work surface in the hood. Aspirate the medium using an aspiration system or electronic pipette, and then wash twice with 5 mL of PBS (room temperature).
- Move the pre-heated trypsin to the work surface, and aspirate the final PBS wash; then, add the pre-heated trypsin to detach the cells (1 mL for a T75 culture flask). Place the culture flask back into the incubator for 2-3 min at 37°C.
- Move the pre-heated medium to the work surface toward the end of the incubation. Verify that cells have detached using a brightfield microscope.
- If all the cells have not detached, apply several careful but firm taps to the side of the flask to detach the remaining cells.
- Return the culture flask to the work surface. Add 4 mL of cell culture medium to the culture flask using an electronic pipette to stop the trypsin action. When adding culture medium to the flask, use the pipette to repeatedly disperse the medium across the surface to detach and gather the cells into a cell suspension.
- Pipette the cell suspension (5 mL) from the flask into a 15 mL tube.
- Centrifuge the cells at 200 x g for 5 min. Open the tube in the hood, remove the supernatant by aspiration, and resuspend the cell pellet gently in 5 mL of cell culture media.
- Assess the number of cells by analyzing a small volume of the cell suspension in an automated cell counter. Note the number of live cells per milliliter of culture media.
- Move the desired volume (e.g., 150 µL per well) of the cell suspension, calculated for a seeding density of approximately 2 x 104 cells/cm2, into a pre-labeled 15 mL centrifuge tube. Add the pre-heated cell culture medium to the 15 mL centrifuge tube at a pre-calculated volume based on the number of wells. For 12-well plates, use a total volume of 1 mL for each well.
- Ensure that the diluted cell suspension is properly mixed by pipetting the contents of the centrifuge tube up and down several times before dispensing the appropriate volume to each well. Once the cell suspension is dispensed in the well, shake the plate in a carefully controlled manner in each direction to better distribute the cells throughout the wells. Place the 12-well plate into the incubator at 37°C until the next day.
- Check on the cells with a brightfield microscope to evaluate the growth. If the cells have achieved sufficient growth to approximately 80% confluency, proceed to the next steps; otherwise, repeat the evaluation daily until sufficient growth is reached.
2. Experimental procedure
- Calculate the amounts needed of the materials for the experiment according to the stock solution concentrations. Thaw the frozen materials (30 mM CCCP stock solution) at 37 °C, and set the cell culture medium to pre-heat at 37 °C.
- Once thawed, create a working solution of 10 µM CCCP by diluting the stock solution (1:3,000) in cell culture medium.
NOTE: Do not pipette volumes less than 1 µL. - Start the experimental treatments once all the necessary materials are prepared and pre-heated. Aspirate the cell culture medium from the wells of the 12-well plate, and then quickly apply the fresh pre-heated medium to the control wells and the pre-heated medium with 10 µM CCCP solution to the test condition wells.
- Incubate the 12-well plate in the 37°C cell incubator for 2 h. During the incubation period, prepare the fixation solution.
- For the preparation of the fixation solution, use pre-made 4% paraformaldehyde (PFA) to dilute 25% glutaraldehyde (GA) stock solution to 0.2% (1:125). Set the solution to pre-heat at 37°C. For a 12-well plate, 500 µL is sufficient per well.
CAUTION: PFA and GA are toxic chemicals. Work in a chemical hood with protective gear. See their respective SDS for details. - When the incubation period is complete, remove the 12-well plate from the incubator, and place it on the work surface.
NOTE: The work no longer requires a sterile environment. - Aspirate the cell culture medium from the wells, and apply the pre-heated fixation solution. Return the 12-well plate to the 37°C incubator for 20 min.
- Aspirate the fixation solution upon completing the incubation, and wash each well twice with room temperature PBS. It is possible to pause the experiment at this stage of the protocol and continue later.
- If the experiment is paused, add 1 mL of PBS per well, seal the 12-well plate with plastic film (parafilm), and store at 4°C.
3. Staining and mounting of the cells on the coverslips
- Thaw DAPI (nuclear stain) stock, and spin down in a mini centrifuge before opening. Prepare DAPI staining solution by diluting DAPI stock in PBS (1:1,000).
- Aspirate the PBS from the 12-well plate, and apply 1 mL of DAPI staining solution to each well. Incubate in the dark at room temperature for 5 min.
- Aspirate the DAPI staining solution. Wash twice with 2 mL of PBS per well.
- Prepare frosted glass microscope slides (glass slides) by washing them in 70% ethanol, followed by three washes in PBS. Carefully dry off the slides using lint-free paper towels, and direct them toward the light to check for signs of dust or grease.
NOTE: Gloves are required for this step. - Label the glass slides with the experimental details. Transfer the mounting medium to a microcentrifuge tube, and spin down in a mini centrifuge.
- Prepare for mounting the coverslips by arranging the workspace. Keep a 12-well plate with coverslips, labeled glass slides, mounting medium, a pipette, 10 µL pipette tips, lint-free paper towels, and tweezers at the ready.
- Apply 10 µL of mounting medium (ProLong Glass) to a prepared glass slide to mount a coverslip.
- Pick up the coverslip from the 12-well plate using tweezers, and dab moisture off the coverslip by briefly touching the edge and back of the coverslip to the prepared lint-free paper towel. Gently lower the coverslip down onto the droplet of mounting medium.
- Repeat the above two steps for each coverslip. Place the mounting medium droplets to allow for between one and four coverslips per glass slide.
- Ensure the glass slides are on a flat surface to avoid the mounted coverslips moving. Place the glass slides in a dark location at room temperature overnight to allow the mounting medium to set. The samples are now ready for imaging. The experiment can be paused at this stage of the protocol and continued later.
- If the experiment is paused at this stage, then after having allowed the samples to set overnight at room temperature, cover them in aluminum foil to protect them from light, and store them at 4°C.
4. Microscopy and imaging
- Cover the samples in aluminum foil for transport (if not already done).
- Upon arrival at the microscopy facility, use double-distilled H2O with microscope filter paper to clean the PBS residues off the coverslips on the glass slides. Check that there are no spots on the coverslips by holding the glass slides toward a bright light.
- Go through the startup procedure for the microscope. Select the appropriate objective (Plan-Apochromat 63x/1.40 Oil M27), and add immersion medium.
- Place the sample in the sample holder. Within the microscope software, use the "Locate" tab to activate the EGFP fluorescence illumination, and using the oculars, manually adjust the z-level to have the sample in focus. Turn off the fluorescence illumination upon finding the focus.
- Switch to the "Acquire" tab within the microscope software. Use the "Smart Setup" to select the fluorescence channels to be used for imaging. For this experiment, the EGFP and DAPI channel presets were selected.
- Adjust each channel intensity from the initial settings using the intensity histogram as a guide for optimized signal strength. Imaging can now begin.
- For imaging, use the software's option to have the imaging positions placed in an array, and center the array in the middle of the coverslip with 12 total positions to be imaged. Verify that each position in the array contains cells. If there are no cells, adjust the position to an area with cells.
- Adjust the focus of each position in the array using the microscope software's autofocus, and follow this with a manual fine adjustment to ensure as many mitochondria as possible are in focus.
NOTE: The EGFP channel is used for these manual adjustments. - Obtain the images using this method for each coverslip. Save the image files, and proceed to the steps for morphological analysis.
5. Generating simulated training data
- Download the code10, and unzip the contents. Follow the instructions in README.md to set up the required environment.
- Navigate to the folder named "src", which is the home folder for this project. The numbered folders inside contain codes that are specific to different steps of using the tool.
NOTE: Use the command "cd <<path>>" to navigate to any subfolders of the code. To run any python file, use the command, "python <<file name>>.py". The presentation named "Tutorial.pptx" contains a complete set of instructions for using the segmentation model. - Make a copy or use the folder "2. Mitochondria Simulation Airy", and rename it (Airy is used here as it is the PSF function closest to a confocal microscope, which was used as the current microscope). Go into the folder named "simulator".
NOTE: This folder contains all the files related to the simulation of the training data. There are three sets of parameters to be set for the simulation. - First, for the simulator in the batch config file "simulator/batch/bxx.csv", set the parameters for about the sample, including the number of mitochondria, the range of diameters and the lengths of the structures, the range of the z-axis that the structure exhibits, and the density of the fluorophores.
- Next, set the parameters related to the optical system.
- This set includes the type of microscope (which determines which PSF model is selected), the numerical aperture (N.A.), the magnification (M), the pixel size (in µm), the emission wavelength of the fluorophores, and the background noise parameter, etc.
- Set the optical parameters of N.A., the magnification, and the minimum wavelength of the dataset in the file "simulator/microscPSFmod.py".
- Set the desired value for the pixel size, and set the emission wavelength of the dataset as a parameter to the "process_matrix_all_z" function in the file "simulator/generate_batch_parallel.py".
- Set the last three parameters of the function "save_physics_gt" in the file "simulator/generate_batch_parallel.py". The parameters are pixel size (in nm), size of the output image, and max_xy.
- Set the third set of parameters regarding the output dataset, such as the size of the output images, the number of tiles in each image, and the number of total images, in the file "simulator/generate_batch_parallel.py".
- Run the file "simulator/generate_batch_parallel.py" to start the simulation.
- To obtain the final-size image, make a copy of the folder named "5. Data Preparation and Training/data preparation" in the home folder, and navigate into it.
NOTE: Each image of the synthetic dataset is formed by creating a montage of four simulated images of 128 pixels x 128 pixels, which gives a final image size of 256 pixels x 256 pixels. This first generates many individual tiles (around 12,000) for both the microscope images (in the "output" folder) and ground truth segmentations (in the "output/physics_gt" folder).- Set the parameters of the batch number, the number of images per batch, and the range of noise in "data_generator.py".
- Run the file "data_generator.py" to create the montage images.
- Copy the folders named "image" and "segment" into the "5. Data Preparation and Training/datatrain/train" folder from the folder "5. Data Preparation and Training/data preparation/data".
6. Deep learning-based segmentation
- Train the segmentation model on the simulated images as follows:
- For training the segmentation model for a new microscope, navigate to the "5. Data Preparation and Training/train" folder, and set the parameters of the batch size, the backbone model for the segmentation, the number of epochs, and the learning rate for training in the file "train_UNet.py".
- Run "train_UNet.py" to start the training. The training process displays the metric for the performance of the segmentation on the simulated validation set.
NOTE: After the training is complete, the model is saved as "best_model.h5" in the "5. Data Preparation and Training/train" folder.
- Test the model on real microscope images that are split to a size that is desirable for the trained model through the following steps.
- Navigate to "6. Prepare Test Data", and copy the ".png" format files of the data into the folder "png".
- Run the file "split_1024_256.py" to split the images to a size that is desirable for the trained model. This creates 256 pixel x 256 pixel sized crops of the images in the "data" folder.
- Copy the created "data" folder into the "7. Test Segmentation" folder.
- Navigate to the folder "7. Test Segmentation", and set the name of the saved model to be used.
- To segment the crops, run the file "segment.py". The segmented images are saved into the "output" folder.
7. Morphological analysis: Analysis of the mitochondrial morphology of the two data groups, "Glucose" and "CCCP"
- Arrange the data to be analyzed (one folder for every image, with each folder containing the segmented output crops of one image).
- Download and place the supplementary file named "make_montage.py" in the folder named "7. Test Segmentation".
- Run the file "make_montage.py" to stitch the segmented output back to the original size of the image.
- Create a new folder named "9. Morphological Analysis" inside the "src" folder.
- Install the Skan16 and Seaborn Python packages into the environment using the command "pip install seaborn[stats] skan".
NOTE: The segmentation masks are skeletonized using the library named Skan to enable the analysis of the topology of each individual mitochondrion. - Place the supplementary file "analyze_mitochondria.py" into the folder "9. Morphological Analysis".
- Arrange the images of different groups of the experiment into different folders inside the folder "7. Test Segmentation".
- Set the parameters of "pixel size" and "input path" in the file "analyze_mitochondria.py".
- Run the file "analyze_mitochondria.py" to run the code to skeletonize and create plots of the analysis.
Results
The results from the deep learning segmentations of mitochondria in confocal images of fixed cardiomyoblasts expressing fluorescent mitochondria markers showcase the utility of this method. This method is compatible with other cell types and microscopy systems, requiring only retraining.
Galactose-adapted H9c2 cardiomyoblasts with fluorescent mitochondria were treated with or without CCCP for 2 h. The cells were then fixed, stained with a nuclear dye, and mounted on glass slides for fluorescence microscopy analysis. A confocal microscope was utilized to acquire images of both the control and CCCP-treated cells. We performed our analysis on 12 confocal images, with approximately 60 cells per condition. The morphologic state of the mitochondria in each image was then determined and quantified. The segmentation masks obtained from the trained model were skeletonized to enable the analysis of the topology of each individual mitochondrion for this experiment. The branch length of the individual mitochondria was used as the parameter for classification. The individual mitochondria were classified into morphological classes by the following rule. Specifically, any mitochondrial skeleton with a length less than 1,500 nm was considered a dot, and the longer mitochondria were further categorized into network or rod. If there was at least one junction where two or more branches intersected, this was defined as a network; otherwise, the mitochondrion was classified as a rod. An example image with the mitochondrial skeletons labeled with the morphology classes is shown in Figure 3.
The mitochondrial morphology categorization in Figure 4A shows that it is possible to detect significant changes when CCCP is applied for 2 h; this is most clearly demonstrated by the increase in dots for the CCCP-treated cells.
The mean branch length in Figure 4B is another avenue for illustrating detectable and significant changes in morphology. Both rods and networks were, as expected, significantly reduced relative to control when the cells were treated with CCCP. The significant increase in the mean branch lengths of dots was also expected given the swelling that mitochondria undergo when exposed to CCCP.
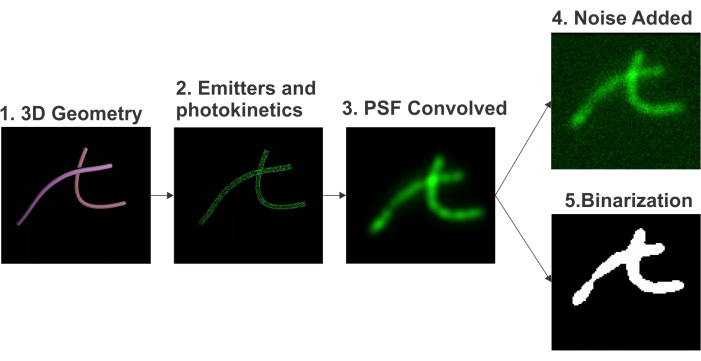
Figure 1: Pipeline for the simulation of fluorescence microscopy images. The pipeline includes (i) 3D geometry generation, (ii) emitters and photokinetics emulation, (iii) 3D PSF convolution, (iv) noise addition, and (v) binarization. Please click here to view a larger version of this figure.
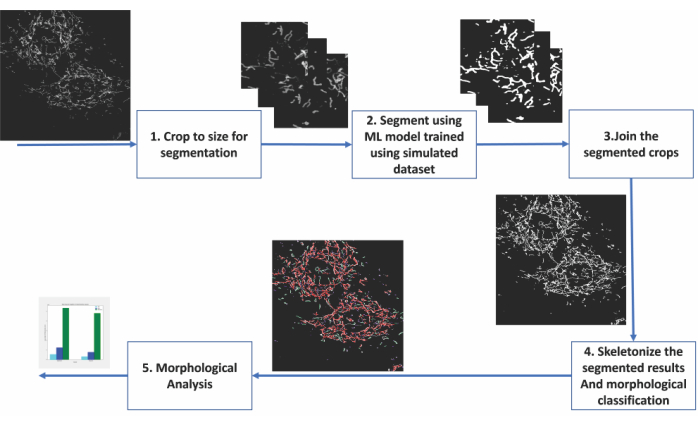
Figure 2: Steps of machine learning-based analysis of mitochondrial morphology. (1) The images to be segmented are first cropped into sizes acceptable for the segmentation model. (2) The deep learning-based segmentation is applied to the image crops. (3) The segmented output crops are stitched back to their original size. (4) The montaged segmentations are skeletonized. (6) Morphological analysis is conducted based on the topology from the skeletonizations. Please click here to view a larger version of this figure.

Figure 3: Mitochondrial skeleton overlayed on the segmentation output of microscopy images. (A) The segmentation output. (B) The straight-line skeleton (Euclidean distance between the start and end points of a branch) is overlayed on top of the segmentation output. The color coding of the skeleton depicts the class of mitochondria. Networks are red, rods are green, and dots are purple in color. Please click here to view a larger version of this figure.
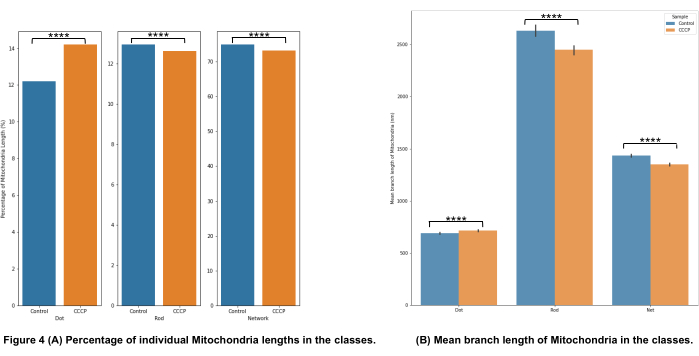
Figure 4: Analysis of mitochondrial morphology. (A) An overview of the relative percentage of different morphological categories based on the total mitochondrial length. (B) Comparison of the mean mitochondrial branch length between the experimental conditions and between morphological categories. The x-axis displays the morphological categorizations, and the y-axis displays the mitochondria mean branch length in nanometers (nm). Statistical significance in the form of p-values is shown as * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. Please click here to view a larger version of this figure.
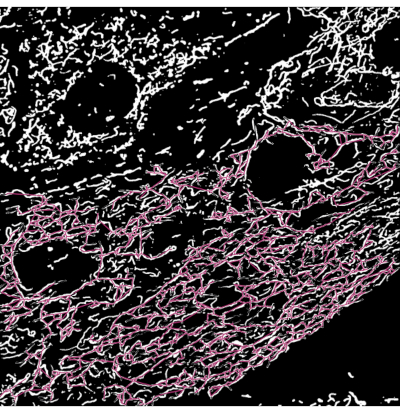
Figure 5: Failure case of segmentation. A high density of mitochondria is a challenging scenario for the segmentation model. The colored skeleton shows the longest single mitochondria detected in the image. By going through the length measurements, these scenarios can be detected and worked on to improve the segmentation results using the morphological operator erode (slims the detected skeleton). Please click here to view a larger version of this figure.
Supplementary Files. Please click here to download the files.
Discussion
We discuss precautions related to the critical steps in the protocol in the paragraphs on "geometry generation" and "simulator parameters". The paragraph titled "transfer learning" discusses modifications for higher throughput when adapting to multiple microscopes. The paragraphs on "particle analysis" and "generating other subcellular structures" refer to future applications of this method. The paragraph on the "difference from biological truth" discusses the different reasons why the simulations could differ from real data and whether these reasons impact our application. Finally, we discuss a challenging scenario for our method in the paragraph "densely packed structures".
Geometry generation
To generate the 3D geometry of mitochondria, a simple 2D structure created from b-spline curves as skeletons works well for the creation of the synthetic dataset. These synthetic shapes closely emulate the shapes of mitochondria observed in 2D cell cultures. However, in the case of 3D tissue such as heart tissue, the shape and arrangement of mitochondria are quite different. In such cases, the performance of the segmentation model may improve with the addition of directionality in the simulated images.
Simulator parameters
Caution should be exercised when setting the parameters of the simulator to ensure they match those of the data to run the inference on, as failure to do this can lead to lower performance on segmentation. One such parameter is the signal-to-noise ratio (SNR) range. The range of the SNR of the data to be tested should match the values of the simulated dataset. Additionally, the PSF used should match that of the target test data. For example, model-trained images from a confocal PSF should not be used to test images from an epi-fluorescent microscope. Another parameter to pay attention to is the use of additional magnification in the test data. If additional magnification has been used in the test data, the simulator should also be set appropriately.
Transfer learning
Transfer learning is the phenomenon of leveraging a learned model trained on one task for use in another task. This phenomenon is also applicable to our problem in relation to different types of microscope data. The weights of the segmentation model (provided with the source code) that is trained on one type of microscopy data can be used to initialize a segmentation model to be used on another kind of optical microscope data. This allows us to train on a significantly smaller subset of the training dataset (3,000 images compared to 10,000), thereby reducing the computational costs of simulation.
Particle analysis
Particle analysis can also be performed on the segmented masks. This can provide information on the area and curvature, etc., of the individual mitochondria. This information can also serve as metrics for the quantitative comparison of mitochondria (not used for this experiment). For example, at present, we define the dot morphology using a threshold based on the length of the mitochondria. It may be useful, in some cases, to incorporate ellipticity to better separate small rod-like mitochondria from puncta or dot-like mitochondria. Alternatively, if certain biological conditions cause the mitochondria to curl up, then curvature quantification may be of interest to analyze the mitochondrial population.
Generating other sub-cellular structures
The physics-based segmentation of subcellular structures has been demonstrated for mitochondria and vesicles1. Although vesicles exhibit varying shapes, their sizes are smaller, and they appear as simple spheres when observed through a fluorescence microscope. Hence, the geometry of vesicles is simulated using spherical structures of an appropriate diameter range. This implies a change in the function generates the geometry (cylinders in the case of mitochondria and spheres in the case of vesicles) of the structures and the respective parameters (step 5.4 in the protocol section). Geometrically, the endoplasmic reticulum and microtubules have also been simulated as tubular structures17. Modeling the endoplasmic reticulum with a 150 nm diameter and microtubules with an average outer diameter of 25 nm and an inner hollow tube of 15 nm in diameter provides approximations of the shapes of these structures. Another parameter that will vary for each of these sub-cellular structures is the fluorophore density. This is calculated based on the distribution of the biomolecule to which the fluorophores bind and the probability of binding.
Difference from biological ground truth
The simulated data used for the simulation-supervised training of the deep learning model differ from real data in many ways. (i) The absence of non-specific labeling in the simulated data differs from real data, as there are often free-floating fluorophores in the real data. This causes a higher average background value in the real image. This difference is mitigated by matching the SNR and setting the background value such that it matches the observed real values. (ii) The motion of subcellular structures and photokinetics are two sources of dynamics in the system. Moving structures in live cells (which move in the range of a few milliseconds) cause motion blur. The exposure time is reduced during real data acquisition to avoid the blurring effect. On the other hand, we do not simulate timelapse and assume motionless structures; this assumption is valid when the exposure time in the real data is small. However, this assumption may produce an error in the output if the exposure time of the real data is large enough to introduce motion blur. Photokinetics, on the other hand, is in the order of nanoseconds to microseconds and can be omitted in the simulation, since the usual exposure times of experiments are long enough (in the order of milliseconds) to average the effects of photokinetics. (iii) Noise in microscope images has different sources, and these sources have different probability density functions. Instead of modeling these individual sources of noise, we approximate it as a Gaussian noise over a constant background. This difference does not significantly alter the data distribution for the conditions of low signal-to-background ratio (in the range of 2-4) and when dealing with the macro density of fluorophores1. (iv) Artifacts in imaging can arise from aberrations, drift, and systematic blurs. We assume the microscope to be well aligned and that the regions chosen for analysis in the real data are devoid of these artifacts. There is also the possibility of modeling some of these artifacts in the PSF18,19,20.
Densely packed structures
The difficulties with not being able to differentiate overlapping rods from networks is a persistent problem in the segmentation of 2D microscopy data. An extremely challenging scenario is presented in Figure 5, where the mitochondria are densely packed, which leads to sub-optimal results in the segmentation model and the following analysis. Despite this challenge, using morphological operators in such situations to slim the skeletonization can help to break these overly connected networks while allowing significant changes in all the mitochondrial morphology categories to still be detected. Additionally, the use of a confocal rather than a widefield microscope for imaging is one method to partially mitigate this problem by eliminating out-of-focus light. Further, in the future, it would be useful to perform 3D segmentation to differentiate mitochondria that intersect (i.e., physically form a network) from rod-like mitochondria whose projections in a single plane overlap with each other.
Deep-learning segmentation is a promising tool that offers to expand the analysis capabilities of microscopy users, thus opening the possibility of automated analysis of complex data and large quantitative datasets, which would have previously been unmanageable.
Disclosures
The authors declare that there are no conflicts of interest related to this article.
Acknowledgements
The authors acknowledge the discussion with Arif Ahmed Sekh. Zambarlal Bhujabal is acknowledged for helping with constructing the stable H9c2 cells. We acknowledge the following funding: ERC starting grant no. 804233 (to K.A.), Researcher Project for Scientific Renewal grant no. 325741 (to D.K.P.), Northern Norway Regional Health Authority grant no. HNF1449-19 (Å.B.B.), and UiT's thematic funding project VirtualStain with Cristin Project ID 2061348 (D.K.P., Å.B.B., K.A., and A.H.).
Materials
| Name | Company | Catalog Number | Comments |
| 12-well plate | FALCON | 353043 | |
| Aqueous Glutaraldehyde EM Grade 25% | Electron Microscopy Sciences | 16200 | |
| Axio Vert.A1 | Zeiss | Brightfield microscope | |
| CCCP | Sigma-Aldrich | C2759 | |
| Computer | n/a | n/a | Must be running Linux/Windows Operating System having an NVIDIA GPU with at least 4GB of memory |
| Coverslips | VWR | 631-0150 | |
| DAPI (stain) | Sigma-Aldrich | D9542 | |
| DMEM | gibco | 11966-025 | |
| Fetal Bovine Serum | Sigma-Aldrich | F7524 | |
| Glass Slides (frosted edge) | epredia | AA00000112E01MNZ10 | |
| H9c2 mCherry-EGFP-OMP25 | In-house stable cell line derived from purchased cell line | ||
| Incubator | Thermo Fisher Scientific | 51033557 | |
| LSM 800 | Zeiss | Confocal Microscope | |
| Mounting Media (Glass) | Thermo Fisher Scientific | P36980 | |
| Paraformaldehyde Solution, 4% in PBS | Thermo Fisher Scientific | J19943-K2 | |
| Plan-Apochromat 63x oil (M27) objective with an NA of 1.4 | Zeiss | 420782-9900-000 | |
| Sterile laminar flow hood | Labogene | SCANLAF MARS | |
| Trypsin | Sigma-Aldrich | T4049 | |
| Vacusafe aspiration system | VACUUBRAND | 20727400 | |
| ZEN 2.6 | Zeiss |
References
- Sekh, A. A., et al. Physics-based machine learning for subcellular segmentation in living cells. Nature Machine Intelligence. 3, 1071-1080 (2021).
- Pernas, L., Scorrano, L. Mito-morphosis: Mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annual Review of Physiology. 78, 505-531 (2016).
- Li, Y., et al. Imaging of macrophage mitochondria dynamics in vivo reveals cellular activation phenotype for diagnosis. Theranostics. 10 (7), 2897-2917 (2020).
- Xu, X., Xu, S., Jin, L., Song, E. Characteristic analysis of Otsu threshold and its applications. Pattern Recognition Letters. 32 (7), 956-961 (2011).
- MorphoLibJ: MorphoLibJ v1.2.0. Zenodo Available from: https://zenodo.org/record/50694#.Y8qfo3bP23A (2016)
- Trainable_Segmentation: Release v3.1.2. Zenodo Available from: https://zenodo.org/record/59290#.Y8qf13bP23A (2016)
- Chu, C. H., Tseng, W. W., Hsu, C. M., Wei, A. C. Image analysis of the mitochondrial network morphology with applications in cancer research. Frontiers in Physics. 10, 289 (2022).
- Xue, F., Li, J., Blu, T. Fast and accurate three-dimensional point spread function computation for fluorescence microscopy. Journal of the Optical Society of America A. 34 (6), 1029-1034 (2017).
- Lanni, F., Gibson, S. F. Diffraction by a circular aperture as a model for three-dimensional optical microscopy. Journal of the Optical Society of America A. 6 (9), 1357-1367 (1989).
- Sekh, A. A., et al. Physics based machine learning for sub-cellular segmentation in living cells. Nature Machine Intelligence. 3, 1071-1080 (2021).
- Liu, Y., Song, X. D., Liu, W., Zhang, T. Y., Zuo, J. Glucose deprivation induces mitochondrial dysfunction and oxidative stress in PC12 cell line. Journal of Cellular and Molecular Medicine. 7 (1), 49-56 (2003).
- Dott, W., Mistry, P., Wright, J., Cain, K., Herbert, K. E. Modulation of mitochondrial bioenergetics in a skeletal muscle cell line model of mitochondrial toxicity. Redox Biology. 2, 224-233 (2014).
- Ishihara, N., Jofuku, A., Eura, Y., Mihara, K. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochemical and Biophysical Research Communications. 301 (4), 891-898 (2003).
- Miyazono, Y., et al. Uncoupled mitochondria quickly shorten along their long axis to form indented spheroids, instead of rings, in a fission-independent manner. Scientific Reports. 8, 350 (2018).
- Ganote, C. E., Armstrong, S. C. Effects of CCCP-induced mitochondrial uncoupling and cyclosporin A on cell volume, cell injury and preconditioning protection of isolated rabbit cardiomyocytes. Journal of Molecular and Cellular Cardiology. 35 (7), 749-759 (2003).
- Nunez-Iglesias, J., Blanch, A. J., Looker, O., Dixon, M. W., Tilley, L. A new Python library to analyse skeleton images confirms malaria parasite remodelling of the red blood cell membrane skeleton. PeerJ. 2018, 4312 (2018).
- Sage, D., et al. Super-resolution fight club: Assessment of 2D and 3D single-molecule localization microscopy software. Nature Methods. 16, 387-395 (2019).
- Yin, Z., Kanade, T., Chen, M. Understanding the phase contrast optics to restore artifact-free microscopy images for segmentation. Medical Image Analysis. 16 (5), 1047-1062 (2012).
- Malm, P., Brun, A., Bengtsson, E. Simulation of bright-field microscopy images depicting pap-smear specimen. Cytometry. Part A. 87 (3), 212-226 (2015).
- Bifano, T., Ünlü, S., Lu, Y., Goldberg, B. Aberration compensation in aplanatic solid immersion lens microscopy. Optical Express. 21 (23), 28189-28197 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved