Method Article
An In Vitro Assay to Study Platelet Migration Using RGD-Functionalized Avidin-Biotin Tethers
In This Article
Summary
A detailed protocol for imaging single migrating platelets using RGD-functionalized avidin-biotin tethers with tunable density is provided, revealing that platelets generate enough force to rupture the avidin-biotin bond.
Abstract
Despite being anucleated cell fragments, platelets are now widely recognized for their multifaceted abilities. Not only do they form blood clots to prevent bleeding after injury, but they also fight infections and maintain vascular integrity during inflammatory diseases. While hemostatic plugs require the collective activation and aggregation of platelets, their role in protecting inflamed blood vessels is performed at the single-cell level. In this context, recent data have shown that platelets can migrate autonomously, a process dependent on the mechanosensing of their adhesive environment. Here, a detailed protocol for imaging single platelet migration is presented, utilizing a three-layer coating system consisting of a poly-L-lysine graft poly(ethylene glycol) (PLL-PEG)-biotin backbone (1), a fluorescent avidin linker (2), and biotin-cyclic Arg-Gly-Asp (cRGD) (3) as the platelet integrin-binding motif. This reductionist approach allows precise control of substrate adhesion properties and serves as a simple, standardized in vitro assay to study the mechanisms underlying platelet migration. The results indicate that migrating platelets binding to cRGD exert forces capable of disrupting the avidin-biotin bond. Furthermore, the density of biotin-cRGD significantly influences both platelet spreading and migration.
Introduction
Platelets are small anucleated cell fragments derived from megakaryocytes in the bone marrow. Activated platelets assemble into aggregates that form the basis of a blood clot to seal vascular lesions in physiological hemostasis or to occlude diseased blood vessels in pathological thrombosis1. In recent decades, however, it has become clear that platelets also play a central role in the pathophysiology of inflammation, infection, and malignancy, where multifaceted interactions with immune cells, cancer cells, and invading pathogens are critical for shaping the host immune response2,3. Recent data have shown that platelets have the ability to migrate autonomously4. In response to systemic bacteremia, platelets are recruited to the liver sinusoids, where they adhere and migrate. Migration allows platelets to capture and bind bacteria, which in turn supports leukocyte activation. Notably, platelets remain intravascular during this process and do not migrate to the sub-endothelial tissue4. In addition, platelets are sentinels of vascular inflammation, scanning inflamed blood vessels for micro-injuries caused by extravasating immune cells5 and employing migration to safeguard inflamed blood vessels6,7. Here, platelets use their integrins to bind fibrinogen deposited on the inflamed endothelium. Platelets then form lamellipodial protrusions that allow them to scan the adhesive properties of their environment. Platelets then polarize and migrate towards higher densities of fibrinogen, a process known as haptotaxis. Directed platelet migration ultimately supports positioning at endothelial microlesions that prevent bleeding in inflamed tissues, including muscle and lung.
Platelet migration is critically dependent on the mechanical properties of the adhesive substrate4,8. The task of sensing and transducing the mechanical properties of the environment is primarily mediated by integrin receptors, which are transmembrane heterodimeric proteins composed of an α- and β-subunit with the extracellular domain binding to the ligand on the extracellular matrix and the intracellular domain binding to the actin cytoskeleton via adaptor proteins9,10. The most abundant platelet integrin is αIIbβ3, and its major ligand is fibrin(ogen)11. Integrin-ligand engagement triggers signaling events that culminate in the formation of an Arp2/3-driven lamellipodium that forms the leading edge of an adherent platelet7. Adherent platelets instantly probe the mechanical properties of their adhesive environment by pulling on it4. When myosin IIa-dependent pulling forces overcome the mechanical stability of the extracellular matrix (e.g., fibrin(ogen)), platelets mechanically rupture the weakened ligands and use their lamellipodial protrusions to scan the microenvironment for adhesive ligands, establish new substrate bonds, and migrate away from the ligand-depleted environment6. Cell migration driven by non-proteolytic, mechanical ligand depletion is likely to play a role beyond platelet function, as it appears to be a general phenomenon observed in cells capable of mechanically remodeling their adhesive matrix8. The rate of ligand depletion determines migration speed and depends on both ligand stability and ligand density of the substrate8. Platelets at low ligand density are able to mechanically disrupt engineered integrin ligands (cyclic Arg-Gly-Asp (cRGD))6,7 bound to glass coverslips with biotin-avidin tethers having an unbinding force of about 160 piconewtons (pN)12,13,14,15.
Here, these observations are exploited to develop a simple in vitro protocol for generating ligand-labile adhesive surfaces, facilitating the study of platelet migration mechanisms. The adhesive coating outlined in this protocol consists of three essential elements: (1) a PLL-PEG-biotin backbone and (2) a neutravidin-fluorescein isothiocyanate (NA-FITC) linker that facilitates the binding of (3) biotin-cRGD (an adhesive ligand) to the PLL-PEG-biotin backbone (also see Figure 1B). The stability of this construct depends on the strength of the biotin-avidin bond, which can break at either the (A) PLL-PEG-biotin-NA-FITC or (B) FITC-NA-biotin-cRGD interface. In addition, the density of adhesive ligands can be tuned by adapting the ratio of PLL-PEG-biotin to PLL-PEG in the bottom layer. Disruption of bond (A) results in the release of FITC from the coating, leading to a decrease in the fluorescence signal underneath migrating platelets. Together, the coating described in this protocol, therefore, allows precise tunability of mechanical and adhesive substrate properties and provides an easy fluorescent-based readout of platelet migration.
Protocol
Animal experiments conducted in this study were performed in compliance with all relevant ethical regulations for studies involving mice and were approved by the local legislation on the protection of animals (Regierung von Oberbayern, Munich, 190-15, 2015). Female and male C57BL/6 mice, 8-10 weeks old (body weight 20-25 g), were used in this study. The details of the reagents and equipment used are listed in the Table of Materials.
1. Biotin-neutravidin-biotin cRGD coating
- Sonicate glass coverslips (24 mm x 24 mm; # 1.5) within 20% HNO3 for 1 min, followed by sonication within isopropanol, ethanol, and H2O for 1 min. After each sonication, rinse the coverslips in distilled water (ddH2O) extensively and finally dry them in the incubator.
- Treat precleaned coverslips with O2 plasma in a plasma cleaner for 2 min, and then assemble with sticky slides as previously described16. Sticky slides and glass coverslips will form a channel between them (as shown in Figure 1A).
NOTE: Efficient plasma treatment is important. Use O2 as a plasma source. - Fill the channel with 2.5 µL of PLL-PEG-biotin (1 mg/mL) diluted in 97.5 µL of PLL-PEG (1 mg/mL) and incubate for 30 min at room temperature (RT), then wash three times with PBS. PLL-PEG-biotin binds neutravidin in the following step, while the PLL-PEG backbone forms an inert substrate that prevents the unspecific binding of proteins and platelets.
NOTE: The concentration of PLL-PEG-biotin in the coating solutions defines the final density of cRGD ligands. - Add 100 µL of Neutravidin-FITC (25 µg/mL) and incubate for 30 min in the dark at RT, then wash three times with PBS.
NOTE: Adjust Neutravidin-FITC concentration to PLL-PEG-biotin concentration. Increase Neutravidin-FITC concentration if higher PLL-PEG-biotin densities are coated. - Add 100 µL of cyclo [Arg-Gly-Asp-D-Phe-Lys (Biotin-PEG-PEG)] (cRGD-biotin) (0.1-1 µM), incubate for 30 min at RT, and wash three times with PBS. Coverslips are ready to use. A schematic of the coating is depicted in Figure 1B.
2. Mouse platelets isolation from blood
- Prepare modified Tyrode's buffer, which contains 136.9 mM of NaCl, 12.1 mM of NaHCO3, 2.6 mM of KCl, 5.5 mM of glucose, 10 mM of HEPES, and adjust the pH to 7.4 and 6.5.
- Anesthetize the mice (following institutionally approved protocols) by intraperitoneal injection of 0.5 mg/kg fentanyl, 5 mg/kg midazolam, and 0.05 mg/kg medetomidine. After confirming the depth of anesthesia via toe pinch, remove the thoracic skin with scissors.
- Prepare a 2 mL syringe (26 G needle) with 150 µL Acid-Citrate-Dextrose (ACD) as an anticoagulant. Insert the needle between the second and third ribs on the left side of the sternum to draw blood from the heart. Afterwards, euthanize the mouse by cervical dislocation.
NOTE: (1) ACD buffer composition: 85 mM of Sodium citrate tribasic dehydrate and 65 mM of citric acid monohydrate in 111 mM of glucose. (2) Blood drawing should be done smoothly to avoid any clot formation. Around 1 mL of blood can be obtained from 8-10-week-old mice. - Mix the blood with 1 mL of Tyrode's buffer (pH 6.5) in a 5 mL FACS (polystyrene round bottom) tube, and centrifuge at 70 x g for 20 min at RT with the brake switched off.
- After centrifugation, take the upper part (around 1 mL) containing platelet-rich plasma (PRP). Mix with 3 mL of Tyrode's buffer (pH 6.5) and add 100 ng/mL prostacyclin (PGI2) to prevent platelet activation.
- Centrifuge at 1200 x g for 5 min at RT, discard the supernatant, resuspend the pellet in 500 µL of Tyrode's buffer (pH 6.5), and measure platelet counts using a hemocytometer.
3. Mouse platelet migration on biotin-neutravidin-biotin cRGD coating
- Supplement the modified Tyrode's buffer (pH 7.4) with 10 x 103/µL mouse platelets, 1 mM of CaCl2, 2 µM of U46619, and 4 µM of Adenosine 5′-diphosphate sodium salt (ADP), and pipette a total volume of 240 µL into the channels prepared in step 1.
- Record live platelet migration using an inverted microscope equipped with a stage incubator. Alternatively, incubate the channel for 1 h at 37 °C in an incubator.
- Fix the sample with 4% paraformaldehyde for 10 min at RT and wash five times with PBS.
- Permeabilize platelets with Triton-X (0.2% in PBS) for 5 min, and wash them five times with PBS.
- Dilute 2.5 µL of Alexa Fluor 594 phalloidin (40x methanolic stock) in 100 µL of PBS. Incubate with platelets for 30 min at dark, then wash five times with PBS.
- Image the slides under a fluorescence microscope.
4. Quantification
NOTE: Platelets that form filopodia (finger-like protrusions) or lamellipodia (sheets-like protrusions) were counted as adherent platelets4. Platelets with a migrating distance of more than one of its diameter were defined as migrating platelets.
- Fraction of migrating platelets: Count the platelet adhesion or migration numbers with the Multi Point tool in Fiji. Right-click on the drop-down menu of Point Tool in the toolbar to select the Multi-point tool. Calculate the fraction of migrating platelets by dividing the number of migrating platelets by the number of adhesive platelets.
- Mean distance of migration: Extract the distance of migration from fixed samples by measuring the length of the migration path "imprinted" in the neutravidin-FITC-coating using the Free Hand Line Tool. Right-click on the drop-down menu of Straight Line in the toolbar to select the Free Hand Line tool.
- Platelet shape descriptors: Generate binary masks by segmenting fluorescent platelets (Alexa594-Phalloidin) using the Threshold function. Select Image > Adjust > Threshold in the toolbar.
- Shape descriptors such as area, perimeter, circularity, and aspect ratio can be obtained in analyze particles. Select shape descriptors in Analyze > Set measurements, then select Display results in Analyze > Analyze Particles.
Results
Activated platelets readily adhere and spread on PLL-PEG-biotin-neutravidin-FITC-biotin-cRGD coated slides (Figure 1C and Figure 2A; 0 min and 5 min) and subsequently polarize by forming a lamellipodium at the leading edge (Figure 2A; 10 min). During this process, the pseudonucleus (dark area in the center of platelets) moved from the center to the rear of the platelet (Figure 2A). Polarized platelets then start to migrate without obvious directionality (Figure 2A; 15 min and 20 min and Figure 2B). We observed that platelets migrating on PLL-PEG-biotin-neutravidin-FITC-biotin-cRGD coatings can break the neutravidin-FITC-biotin-PLL-PEG bond, as evidenced by the reduction in fluorescence intensity along the path of migration (Figure 1C and Figure 2A,B). Moreover, the disrupted neutravidin-FITC-biotin-cRGD complex accumulates on the platelet surface (Figure 2A,B). This phenomenon is similar to previous findings where migrating platelets removed fibrinogen from the substrate and accumulated it within their open canalicular system (OCS)4.
To investigate how ligand density influences platelet migration, the cRGD density was adjusted by varying the ratio of PLL-PEG-biotin to PLL-PEG in the first coating layer. These data reveal that mouse platelets achieve optimal migration at a 2.5% concentration of PLL-PEG-biotin. Migration is reduced at both lower (1%) and higher (10%) concentrations of PLL-PEG-biotin (Figure 3A,B). These substrate-dependent changes in migratory behavior are accompanied by alterations in platelet morphology (Figure 3C). At low ligand densities (1%), platelets do not spread adequately, as evidenced by a low projected platelet area and perimeter (Figure 3C). This suggests insufficient integrin activation and outside-in signaling7. Consequently, platelets are unable to exert forces on the cRGD ligands, fail to remodel the substrate, and do not migrate (Figure 3A,B).
At an intermediate ligand density (2.5%), there is a significant increase in platelet area and perimeter (Figure 3C). Platelets spread effectively, mechanically disrupting the labile cRGD ligands, and migrate (Figure 3A,B). However, at high ligand densities (10%), although spreading increases, platelets fail to polarize, indicated by a reduced aspect ratio (Figure 3C). Under these conditions, platelets remain attached to the sticky substrate and do not migrate because they cannot break the labile cRGD ligands (Figure 3A,B).
These findings demonstrate that platelet migration is critically dependent on the adhesive properties of the substrate. Platelets can only migrate when they sufficiently engage with adhesive ligands and simultaneously generate traction forces strong enough to overcome the tension tolerance of the adhesive substrate4,6,7,8.
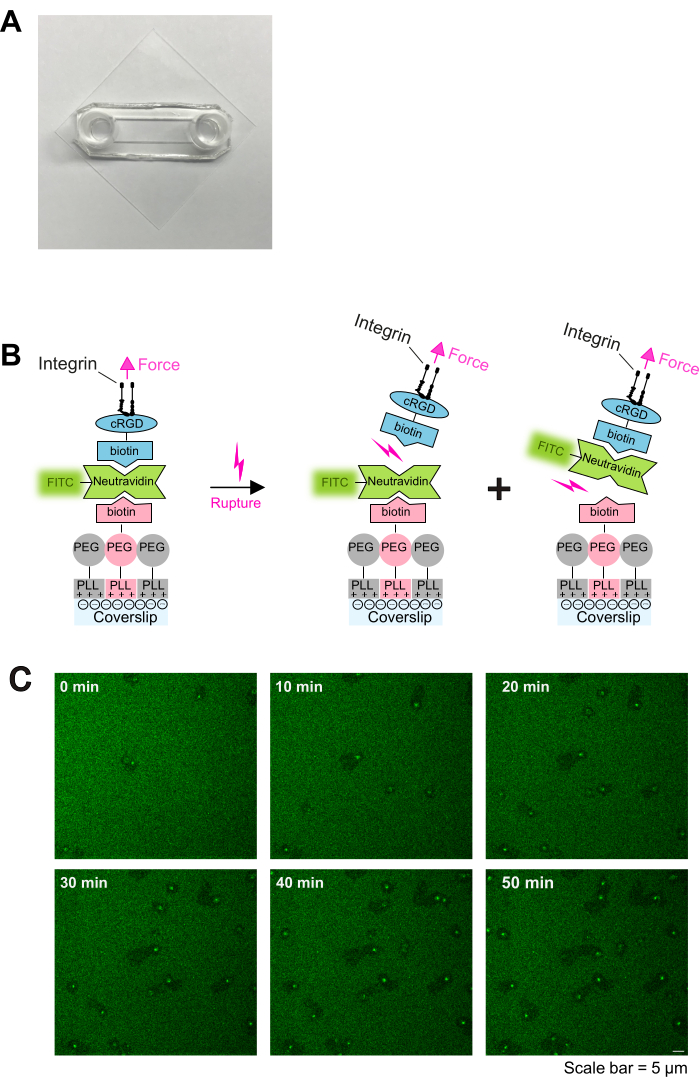
Figure 1: Coating procedure and live imaging of platelet-substrate interaction. (A) Photograph of the assembled chamber. (B) Principle of the protocol. Left: Schematic of the coating. Plasma-treated glass coverslips were coated with three layers. The first layer is the mixture of PLL-PEG and PLL-PEG-biotin (the PLL-PEG backbone is inert to cells and prevents unspecific binding). The second layer is neutravidin-FITC, and the third layer is biotin-cRGD. Neutravidin-FITC bridges PLL-PEG-biotin and biotin-cRGD. Platelets bind to the biotin-cRGD via integrins in the plasma membrane. Right: Illustration of biotin-avidin unbinding by platelet integrins. Platelets exert forces on the substrate through integrin-cRGD engagement and rupture either the upper cRGD-biotin - neutravidin-FITC - bond or the lower neutravidin-FITC - PLL-PEG-biotin - bond. Rupture of the lower bond between neutravidin-FITC and biotin-PLL-PEG results in a reduced fluorescent signal, while rupture of the upper bond between biotin-cRGD and neutravidin-FITC does not. (C) Time series showing platelet migration and depletion of labile cRGD-ligands. Areas of reduced fluorescence indicate the migration tracks of platelets (also see Figure 2A), and areas of increased fluorescence indicate FTIC-neutravidin-biotin-cRGD accumulation on migrating platelets. Scale bar: 5 µm. Please click here to view a larger version of this figure.

Figure 2: Immunofluorescence imaging of platelet migration encoded by the remodeled substrate. (A) Representative movie sequence of single platelet migration. Left: Phase contrast images of migrating platelets. Platelets polarize by forming a lamellipodium at the leading edge and subsequently migrate. Right: Neutravidin-FITC was removed from the substrate, leaving the "footprint" of platelet migration. Ruptured cRGD-biotin-neutravidin-FITC accumulates at the center of migrating platelets. (B) Representative images of platelet migration on ligand labile cRGD-biotin-avidin coatings. Upper: Platelets stained with phalloidin (Alexa FluorTM 594) showed large lamellipodium formation and a polarized shape reminiscent of migrating platelets. Migrating platelets rupture the cRGD-biotin-neutravidin-FITC bond, generating paths of migration on the substrate indicated by reduced FITC signal. Lower: cropped image with larger magnification. (C) Migration tracks were manually drawn in Fiji with yellow lines. Scale bars: 5 µm. Please click here to view a larger version of this figure.

Figure 3: The ratio of PLL-PEG-biotin to PLL-PEG determines platelet migration and shape. (A) Representative images of platelets on 1%, 2.5%, and 10% PLL-PEG-biotin coatings, scale bar: 5µm. (B) Quantification of platelet adhesion, migration efficiency, and migration distance, n = 4 independent experiments; Mean/SD; One-way ANOVA/Tukey; **p < 0.01; ***p < 0.001; ****p < 0.0001, ns: not significant (C) Quantification of platelet shape descriptors (area, perimeter, circularity, and aspect ratio), n = 4 independent experiments; Mean/Min-Max; One-way ANOVA/Tukey; **p < 0.01; ***p < 0.001; ****p < 0.0001, ns: not significant. Please click here to view a larger version of this figure.
Discussion
In this protocol, a three-layer coating procedure is presented consisting of (1) a PLL-PEG-biotin backbone and (2) a neutravidin-FITC linker that facilitates the binding of (3) biotin-cRGD (an adhesive ligand) to the PLL-PEG-biotin backbone (also see Figure 1B) which allows precise tunability of mechanical and adhesive substrate properties by varying the ratio of PLL-PEG-biotin to PLL-PEG and provides an easy fluorescent-based readout of platelet migration. While this protocol uses FITC-conjugated neutravidin and Alexa Fluor 594-conjugated phalloidin to track platelet migration and shape, other fluorophore-conjugated avidin and phalloidin can be used. Since all coating reagents, including PLL-PEG, PLL-PEG-biotin, neutravidin-FITC, and cRGD-biotin, as well as the platelet activators are commercially available, this experiment can be performed without much additional effort. The critical step is the plasma treatment because inefficient plasma treatment leads to inadequate binding of PLL-PEG/PLL-PEG-biotin to the substrate, resulting in failure of platelet adhesion and migration. The performance of the plasma cleaner is important. In general, plasma cleaners with 13.6 MHz generators perform better than those with 40 KHz or 100 KHz. In addition, an oxygen plasma source is more suitable than ambient air. After successful plasma treatment, the coating steps are almost not prone to errors.
Platelets have previously been shown to use the mesenchymal mode of migration, which is highly dependent on substrate adhesions4,17. Analogous to fibroblasts, low ligand densities impede migration as platelets struggle to adhere to the substrate, whereas high ligand densities hinder migration by preventing the disengagement of firm adhesions18 (Figure 3). At intermediate densities, platelets do adhere while myosin IIA-dependent pulling forces per integrin bond are still sufficient to break adhesions. It is noteworthy that the high affinity between integrin αIIbβ3 and its ligand fibrin(ogen) results in the disassembly of adhesions not through the release of the adhesive ligand, but rather through its rupture or detachment from the underlying substrate, such as endothelial cells in vivo or coverslips in vitro4,6. The unbinding force required to detach fibrinogen from the underlying substrate is heavily influenced by physical and (bio-) chemical properties that are often challenging to control, which may lead to significant variability in migration efficiency. The assay introduced here offers a reductionist approach to address this issue. However, the breaking force of the avidin-biotin bond is fixed at around 160 pN, so more sophisticated tension sensors, such as those based on a double-stranded DNA tether, must be used to study platelet adhesion at lower force regimes19.
Previous studies found that platelet migration is involved in bacterial infection and vascular inflammation in vivo. Consequently, platelets with migration defects, such as impaired myosin contractility, actin polymerization, or integrin outside-in signaling, show impaired bacteria clearance and vascular integrity4,6,7. The assay presented here is simple, stable, and, therefore, suitable for screening small molecular inhibitors and their effects on platelet migration. It provides a novel in vitro tool to explore therapeutic targets in inflammation or thrombosis.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) project numbers 514478744 and 514477451 to F.G. The project is funded by the European Union (ERC, MEKanics, 101078110). Views and opinions expressed are those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council Executive Agency. Neither the European Union nor the granting authority can be held responsible for them.
Materials
| Name | Company | Catalog Number | Comments |
| Adenosine 5′-diphosphate sodium salt (ADP) | Sigma-Aldrich | A2754 | |
| Alexa Fluor 594 phalloidin | Thermofisher | A12381 | |
| Blood counter | Sysmex | XN-1000 | |
| Bottomless 6 channel sticky slide | IBIDI, sticky slides VI0.4 | 80608 | |
| Centrifuge | Eppendorf | 5804 | |
| Confocal microscope | Zeiss | LSM880 | |
| cyclo [Arg-Gly-Asp-D-Phe-Lys(Biotin-PEG-PEG)] | Peptide international | PCI-3697-PI | |
| FACS tubes | Corning Brand | 352052 | |
| FITC conjugate neutravidin | Thermofisher | A2662 | |
| Formaldehyde | Thermofisher | 28908 | |
| HEPES solution | Sigma-Aldrich | H0887 | |
| Phase contrast and epifluorescent microscope | Olympus | IX83 | |
| Plasma cleaner | Diener | 116531 | |
| PLL(20)-g[3.5]-PEG(2)/PEG(3.4)-biotin(50%) | Susos | PLL(20)-g[3.5]-PEG(2)/PEG(3.4)-biotin (50%) | |
| Poly(L-lysine)-graft-poly(ethylene glycol) co-polymer | Susos | PLL(20)-g[3.5]-PEG(2) | |
| Prostaglandin I2 sodium salt (PGI2) | Abcam | ab120912 | |
| Sonicator | BANDELIN | SONNOREX RK514M | |
| Ttriton X-100 20% v/v | Cayman Chemical | 600217 | |
| U46619 | Enzo Life Sciences | BML-PG023-0001) |
References
- Jackson, S. P. Arterial thrombosis--insidious, unpredictable and deadly. Nat Med. 17, 1423-1436 (2011).
- Kaiser, R., Escaig, R., Nicolai, L. Hemostasis without clot formation: how platelets guard the vasculature in inflammation, infection, and malignancy. Blood. 142 (17), 1413-1425 (2023).
- Gaertner, F., Massberg, S. Patrolling the vascular borders: Platelets in immunity to infection and cancer. Nat Rev Immunol. 19 (12), 747-760 (2019).
- Gaertner, F., et al. Migrating platelets are mechano-scavengers that collect and bundle bacteria. Cell. 171 (6), 1368-1382.e23 (2017).
- Gros, A., et al. Single platelets seal neutrophil-induced vascular breaches via GPVI during immune-complex-mediated inflammation in mice. Blood. 126 (8), 1017-1026 (2015).
- Nicolai, L., et al. Vascular surveillance by haptotactic blood platelets in inflammation and infection. Nature Comm. 11, 5778 (2020).
- Kaiser, R., et al. Mechanosensing via a GpIIb/Src/14-3-3zeta axis critically regulates platelet migration in vascular inflammation. Blood. 141 (24), 2973-2992 (2023).
- Sarkar, A., LeVine, D. N., Kuzmina, N., Zhao, Y., Wang, X. Cell migration driven by self-generated integrin ligand gradient on ligand-labile surfaces. Curr Biol. 30 (20), 4022-4032 (2020).
- Hynes, R. O. Integrins: Bidirectional, allosteric signaling machines. Cell. 110 (6), 673-687 (2002).
- Kechagia, J. Z., Ivaska, J., Roca-Cusachs, P. Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol. 20, 457-473 (2019).
- Shattil, S. J., Newman, P. J. Integrins: Dynamic scaffolds for adhesion and signaling in platelets. Blood. 104 (6), 1606-1615 (2004).
- Yuan, C., Chen, A., Kolb, P., Moy, V. T. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry. 39 (33), 10219-10223 (2000).
- Florin, E. L., Moy, V. T., Gaub, H. E. Adhesion forces between individual ligand-receptor pairs. Science. 264 (5157), 415-417 (1994).
- Merkel, R., Nassoy, P., Leung, A., Ritchie, K., Evans, E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397, 50-53 (1999).
- Jurchenko, C., Chang, Y., Narui, Y., Zhang, Y., Salaita, K. S. Integrin-generated forces lead to streptavidin-biotin unbinding in cellular adhesions. Biophys J. 106 (7), 1436-1446 (2014).
- Fan, S., Lorenz, M., Massberg, S., Gaertner, F. Platelet migration and bacterial trapping assay underflow. Bio-Protocol. 8 (18), e3018 (2018).
- Lauffenburger, D. A., Horwitz, A. F. Cell migration: A physically integrated molecular process. Cell. 84 (3), 359-369 (1996).
- Palecek, S. P., Loftus, J. C., Ginsberg, M. H., Lauffenburger, D. A., Horwitz, A. F. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 385, 537 (1997).
- Zhao, Y., Wetter, N. M., Wang, X. Imaging integrin tension and cellular force at submicron resolution with an integrative tension sensor. J Vis Exp. (146), e59476 (2019).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved