Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Computer-assisted Large-scale Visualization and Quantification of Pancreatic Islet Mass, Size Distribution and Architecture
W tym Artykule
Podsumowanie
Novel computer-assisted methods of large-scale procurement and analysis of immunohistochemically stained pancreatic specimens are described: (1) Virtual Slice capture of the entire section; (2) Mass analysis of large-scale data; (3) Reconstruction of 2D Virtual Slices; (4) 3D islet mapping; and (5) Mathematical analysis.
Streszczenie
The pancreatic islet is a unique micro-organ composed of several hormone secreting endocrine cells such as beta-cells (insulin), alpha-cells (glucagon), and delta-cells (somatostatin) that are embedded in the exocrine tissues and comprise 1-2% of the entire pancreas. There is a close correlation between body and pancreas weight. Total beta-cell mass also increases proportionately to compensate for the demand for insulin in the body. What escapes this proportionate expansion is the size distribution of islets. Large animals such as humans share similar islet size distributions with mice, suggesting that this micro-organ has a certain size limit to be functional. The inability of large animal pancreata to generate proportionately larger islets is compensated for by an increase in the number of islets and by an increase in the proportion of larger islets in their overall islet size distribution. Furthermore, islets exhibit a striking plasticity in cellular composition and architecture among different species and also within the same species under various pathophysiological conditions. In the present study, we describe novel approaches for the analysis of biological image data in order to facilitate the automation of analytic processes, which allow for the analysis of large and heterogeneous data collections in the study of such dynamic biological processes and complex structures. Such studies have been hampered due to technical difficulties of unbiased sampling and generating large-scale data sets to precisely capture the complexity of biological processes of islet biology. Here we show methods to collect unbiased "representative" data within the limited availability of samples (or to minimize the sample collection) and the standard experimental settings, and to precisely analyze the complex three-dimensional structure of the islet. Computer-assisted automation allows for the collection and analysis of large-scale data sets and also assures unbiased interpretation of the data. Furthermore, the precise quantification of islet size distribution and spatial coordinates (i.e. X, Y, Z-positions) not only leads to an accurate visualization of pancreatic islet structure and composition, but also allows us to identify patterns during development and adaptation to altering conditions through mathematical modeling. The methods developed in this study are applicable to studies of many other systems and organisms as well.
Protokół
1. Creating Virtual Slices of Immunohistochemically Stained Images
- Open StereoInvestigator. Place the clean slide holding the sample in the microscope holder and visualize it in Stereo Investigator by clicking on "Acquisition" and then "Live Image" (Acquisition→ Live Image). Determine exposure levels for each channel; in our specific example, Channel 2 is used for DAPI, Channel 3 for GFP, Channel 4 for RFP, Channel 5 for Cy5, and Channel 6 for Cy7. Use the "Video Histogram" window, which displays the intensity of the fluorescence, to determine the exposure level. Appropriate fluorescence intensities are reached when the intensity tails off at the right end of the Video Histogram. Exposure levels can be changed in the "Camera Settings" window. The Video Histogram can be focused with the microscope focus wheel.
- Before creating a Virtual Slice, contour the sample by clicking on the screen at a point away from the sample, which will mark a reference point, and then clicking around the outline of the sample. When the contour is complete, right click and select "Close Contour" to connect the beginning and end points.
- Once the contour is closed, capture a Virtual Slice for the sample (Acquisition→ Acquire Virtual Slice). When the Virtual Slice window options open, select "High Speed Acquire" as well as automatic focusing by unchecking the box next to "Manual." Save the file.
- The virtual slice prefocus must be calibrated by selecting (right click→ Add to Focus Site List) and manually focusing several random sections, i.e., sites, in the Virtual Slice preview. When several sites have been focused, start the Virtual Slice (right click→ Start Virtual Slice with Prefocus). After the Virtual Slice for the first sample is complete, switch to the next channel and adjust the exposure accordingly. Repeat steps 4 and 5 for each channel.
2. Computer-assisted Two-Dimensional Analysis
Quantification of Islets
- The images are then processed using an ImageJ macro named "IHCVS," which first prepares loaded images for analysis and then quantifies features of islets such as cellular composition (i.e. each area of beta-, alpha-, and delta-cell populations, and islet area at the sum value). The stack is split into its separate channels in ImageJ, where each individual channel shows a separate image based on the immunohistochemical staining. Each image is then automatically thresholded, after which an 8-bit black and white image, or a mask, of the thresholded image is produced. The separate channel images are then added together arithmetically into a composite mask, and ImageJ's built-in particle analysis is performed on the composite image.
- ROIs are then identified while excluding particles smaller than one single beta cell (<170 μm2; area calculated using a diameter of ~15 μm; Hara et al., 2003). The resultant ROIs are analyzed and quantified using ImageJ; quantification can include perimeter (a distance surrounding an area), circularity (a degree of roundness where the number 1.0 depicts a perfect circle), Feret's diameter (the longest distance within an area) and center coordinates for each analyzed region so that the islet distribution can be mathematically analyzed using various combinations of these parameters and plotted in a 3D graph, the data of which will be used for mathematical modeling. A single virtual slice image is analyzed in 30 seconds. Multiple images can be analyzed by embedding the image processing and analysis script into a loop syntax supported by ImageJ macro language. The loop opens all the files in a given directory, analyzes the images, and outputs results into a new location as an Excel file.
- The aggregated results are saved into an Excel spreadsheet, storing data such as area, perimeter, circularity, Feret's diameter, and islet center for each islet, along with corresponding numbers labeled in the image.
Computational Analysis and Histogram Setup
- Mathematica scripts, which run automatically once started, are used to process the spreadsheet data collected, amounting to hundreds of thousands of entries. One of the critical processes that are run is a frequency analysis, which constructs a histogram used to examine size distribution and also to determine outliers in the data. Under the script, total islet area is measured and compared to other parameters, particularly to time points or the age of the pancreas, and then accordingly displayed in a graph.
3. Three-Dimensional Reconstruction of Two Dimensional Immunohistochemically Stained Virtual Slice Images
3D Reconstruction of Virtual Slices
- Copy the script ("im_jp2_2_tiff") into the directory (i.e., folder) that contains the jp2 images. Run the script with the Linux shell by typing "./im_jp2_2_tiff" into the console and then pressing enter. Running this script will automatically convert all of the jp2 images in the directory into tiff files, which is the format used in the construction and quantification of three-dimensional stacks from the Virtual Slices. (Note: Type in "chmod+x im_jp2_2_tiff" if the script does not run, which will convert the text file into a script so that it can run properly.)
- When all of the captured images have been converted from jp2 to tiff files, they are ready to be used for analysis in ImageJ.
- With ImageJ, open all of the images for the first sample (five images in this case: DAPI, GFP, RFP, Cy5, Cy7) and merge them as one image (Image→ Color→ Merge Channels). Repeat this process for each of the samples. When all of the samples have been combined as single images, clean the images by selecting unwanted regions with the "Polygon Selections" tool and filling them in (Edit→ Fill). (Resize the images to a smaller size, if necessary). Then convert each image into an RGB color image (Image→ Type→ RGB color). Finally, create a stack out of the images (Image→ Stacks→ Image to Stacks), after which they can further be aligned with the "Stack Reg" plugin, if necessary. Ultimately, merging the channels and then combining the images creates a 3D montage of all of the islets in the entire pancreas. To view the image, click on "Plugins" and then "3D viewer."
4. Islet Mapping
Collecting Image Stacks
- Situate the whole mount pancreatic islets under the microscope.
- Open the Slidebook software. Access the "Capture and Focus Control" by pressing "Ctrl+Shift+E." In the "Focus Control" window, set the objective to 20X or 40X, depending on the size of the islet. Apply water if using a water immersion objective lens.
- In order to use the microscope eyepiece to locate islets, set "Bin" to 2X and set the Filter Set to User1 in the "Focus Controls" window. "Emission Selection" should be set to 100% Eyes and Neutral Density to 1. Click "GFP ey" and then "Open Fluor."
- To visualize the sample in Slidebook, return to the Focus Controls window and switch "Filter Set" to "Live" and click "GFP ds." Use the joystick to center the islet in the Camera 1 window as well as possible. Focus to a depth somewhere near the center of the islet, where the cells can be discerned easily.
- In the "Focus Controls Window, " select the "Z tab." Click set in the upper left corner of the window to set the current depth as a reference point. Use the scope control to scroll to the uppermost depth of the islet in which features can be discerned clearly and click "Set Top." Scroll to the bottom of the islet and click "Set Bottom." Click "Go" to return to the reference point.
- In the Capture window, set "Bin Factor" to "2X" and set "Filter Set" to "Live." Set the "Capture Type" to "3D" (and only 3D). On the right side of the window, click "Use Top and Bottom Positions." Click "Return to Reference Point After Capture." Set the "Step Size" to "3."
- Beneath the "Filter Set Box," select DAPI dsu, GFP dsu, RFP dsu, and Cy5 Dsu. For each one, click "Find Best" under "Adjust Exposure," then click "Test" to make sure that the selected exposure is suitable. Click "Start" to begin the capture.
- When the capture is completed, adjust the levels as necessary and change the displayed color for RFP to white. Save the slide and go to View→Export→TIFF series. Type in the name of the slide with a dash at the end and save it. Dozens of individual TIFF files are created, so it may be helpful to keep each islet's image stack in a separate folder.
Mapping Image Stacks
- Open Stereo Investigator. Visualize the image by opening the image stacks (File→ Image Stack → Image Stack Open). In the "Image Scaling menu," set "Distance Between Images" to 3.00 μm. To correct for 2X binning in Slidebook, select "Override X and Y Scaling" and "User Specified for Source of X and Y" Scaling, and enter 0.65 for both X and Y.
- Center the image by clicking in the middle of the islet of interest in the "Macro window." Mark at least one cell using the appropriate marker. Beta-cells will appear green, alpha cells will appear red, and delta cells will appear white; use Marker 2 (a hollow circle) to mark beta-cells, Marker 5 (a hollow triangle) to mark alpha cells, and Marker 6 (a hollow triangle) to delta cells. Cells should be marked in the center of their nucleus, which will appear blue if the sample has been stained with DAPI.
- Once at least one cell has been marked, display the "Orthogonal View" using the icon in the top menu. Check "Z-Filter" and "Symmetric." The range should be set to 15.00. Starting at the 0.0 Z level, scroll through the islet using the mouse wheel and mark each cell with the appropriate marker. Each cell should be marked only once at the center of its depth. Upon finishing marking each of the Z-levels, the "Z-filter" checkbox can be unchecked. This will show all of the markers from all of the Z-levels.
- When every cell has been marked, save the file as a "DAT" file, then go to File→ Export Tracing. Save the tracing as a "TXT" file. Click "New Data File" to clear the workspace before attempting to map new islets.
5. Representative Results:
The preparation of Virtual Slices out of an immunohistochemically stained pancreas sample enables one to examine all of the endocrine cells (alpha, beta, and delta-cells) in the whole pancreas, both together as islets (Figure 1A) and individually in separate channels (Figure 1B). With the assistance of computer programs and scripts, a mass analysis of large-scale data can be performed on these Virtual Slices. Specifically, a particle analysis of composite masks (Figure 1C) is output as a statistics table containing such parameters as islet area, perimeter (the distance surrounding an area), circularity (a degree of roundness, where 1.0 represents a perfect circle), and Feret's diameter (the longest distance within an area) for each islet detected (Figure 1D). The large-scale analysis of these images results in the production of total islet number and size distribution histograms as well as a detailed comparison of alpha-, beta-, and delta-cell areas. In addition, each Virtual Slice is taken at a depth of approximately 5 μm, and all of the individual 2D Virtual Slices are further stacked to create a 3D reconstruction of the entire pancreatic sample. Islet mapping demonstrates another instance not only of capturing islets in 3D, but also of detailed computer-assisted analysis. Islet mapping consists of the capture of distinct islets (Figure 2A) and the subsequent marking of alpha-, beta-, and delta-cells at various Z-planes (Figure 2B) to visualize an islet in 3D (Figure 2C, D). Automated mathematical analysis of mapped islets displays their cellular composition and architecture, including cell-cell distances (Figure 2E) and cumulative probabilities of cell-cell distance distributions (Figure 2F).
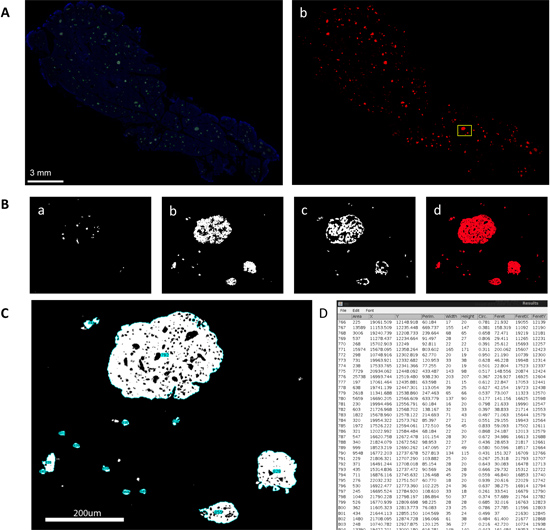
Figure 1. Large-scale capture and analysis of islet distribution using Virtual Slice. A. Virtual slice view of a human pancreatic section. a. Immunohistochemical staining for insulin (green), glucagon (red), somatostatin (white) and DAPI (blue). b. Converted 8-bit mask after automatic thresholding. A boxed area is magnified in B. B. Views of each channel. a. delta-cells, b. beta-cells, c. alpha-cells, and d. merged composite image. C. Particle analysis performed upon composite mask. Note that each islet structure including small cell clusters is numbered (blue highlight). D. Statistics table of various parameters measured for individual structures, which have IDs that correspond to tags shown in C.

Figure 2. Analysis of Immunohistochemical Virtual Slice. A. 3D scatter plot of Figure 1 showing size and shape distribution of each islet by parameters such as area, circularity and feret's diameter. B. 3D scatter plot of Figure 1 showing cellular islet composition and size. C. Islet size distribution of the whole human section analysis from Figure 1 fitted to a lognormal probability density distribution. D. Mathematical analysis of cellular composition ratios (beta-cells in green, alpha-cells in red and delta-cells in blue) for each islet effective diameter bin of Figure 1. E. a. Islet size distribution of random sampling immunohistochemical analysis (left). Islet size distribution of virtual slice analysis (right). b. Log-normal plot comparison of random sampling immunohistochemical analysis (red) and virtual slice (blue).
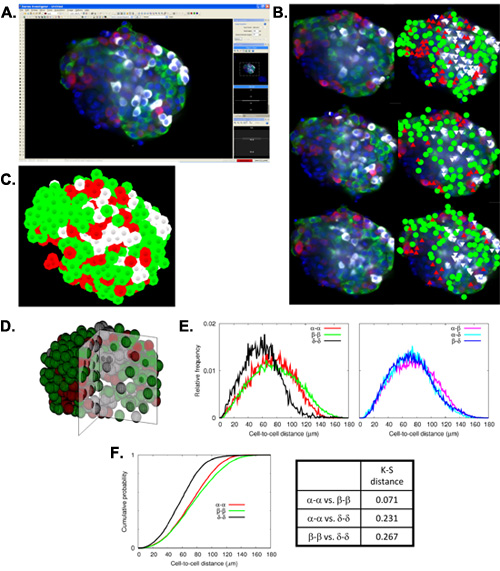
Figure 3. Islet mapping and mathematical analysis of cellular composition and architecture. A: Screen-capture showing a single focal plane from a 3D reconstructed stack of images of a human islet uploaded into Stereo-Investigator (beta-cells in green, alpha-cells in red, delta-cells in white, and nuclei in blue). B: Fluorescence images (left) and corresponding mapped data (right) in three different focal planes shown at an interval of 10 μm. C: Representative view of 3D reconstructed islet mapping data. D: 3D reconstruction of the quarter-sliced islet based on the coordinates obtained by islet mapping. E: Mathematical analysis of cellular composition and architecture. Left. Relative frequency of cell-cell distances between two cells in a single cell population. Right. Relative frequency of cell-cell distances between two different cell populations. F: Kolmogorov-Smirnov (K-S) test. Left. Cumulative probabilities of cell-to-cell distance distributions for alpha-to-alpha, beta-to-beta, and delta-to-delta cells. Right. K-S distances for the corresponding three cumulative probabilities.
Dyskusje
The computer-assisted large-scale visualization and quantification presented here afford four key points in pancreatic islet studies: (1) A large-scale analysis of pancreatic specimens provides a comprehensive view of overall islet size distribution and islet architecture. (2) The 3D reconstruction and mathematical analysis of cellular composition and architecture further facilitate the examination of the spatial arrangement of endocrine cells within an islet. (3) Striking islet plasticity among different species and in ...
Ujawnienia
No conflicts of interest declared.
Podziękowania
The study is supported by US Public Health Service Grant DK-081527, DK-072473 and DK-20595 to the University of Chicago Diabetes Research and Training Center (Animal Models Core), and a gift from the Kovler Family Foundation.
Materiały
| Name | Company | Catalog Number | Comments | |
| Fluorescent microscope | Microscope | Olympus Corporation | IX-81 | |
| Stereo Investigator | Program | MBF Bioscience | ||
| MIP-GFP mice | Mice | Jackson Laboratory | ||
| Mathematica | Program | Wolfram | ||
| Image J | Program | National Institutes of Health | ||
| Slidebook | Program | Olympus |
Odniesienia
- Steiner, D. J., Kim, A., Miller, K., Hara, M. Pancreatic islet plasticity - Interspecies comparison of islet architecture and composition. ISLETS. 2, 135-145 (2010).
- Kim, A., Miller, K., Jo, J., Wojcik, P. l., Kilimnik, G., Hara, M. Islet architecture - a comparative study. ISLETS. 1, 129-136 (2009).
- Kilimnik, G., Kim, A., Jo, J., Miller, K., Hara, M. Quantification of pancreatic islet distribution in situ in mice. Am J Physiol Endocrinol Metab. 297, E1331-E1338 (2009).
- Hara, M., Dizon, R. F., Glick, B. S., Lee, C. S., Kaestner, K. H., Piston, D. W., Bindokas, V. P. Imaging pancreatic beta-cells in the intact pancreas. Am J Physiol Endocrinol Metab. 290, E1041-E1047 (2006).
- Hara, M., Wang, X., Kawamura, T., Bindokas, V. P., Dizon, R. F., Alcoser, S. Y., Magnuson, M. A., Bell, G. I. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am J Physiol Endocrinol Metab. 284, E177-E183 (2003).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone