Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Parallel Interrogation of β-Arrestin2 Recruitment for Ligand Screening on a GPCR-Wide Scale using PRESTO-Tango Assay
W tym Artykule
Podsumowanie
Given that GPCRs are attractive druggable targets, GPCR ligand screening is thus indispensable for the identification of lead compounds and for deorphanization studies. Towards these efforts, we describe PRESTO-Tango, an open-source resource platform used for simultaneous profiling of transient β-arrestin2 recruitment at approximately 300 GPCRs using a TEV-based reporter assay.
Streszczenie
As the largest and most versatile gene superfamily and mediators of a gamut of cellular signaling pathways, G-protein-coupled receptors (GPCRs) represent one of the most promising targets for the pharmaceutical industry. Ergo, the design, implementation, and optimization of GPCR ligand screening assays is crucial, as they represent remote-control tools for drug discovery and for manipulating GPCR pharmacology and outcomes. In the past, G-protein dependent assays typified this area of research, detecting ligand-induced events and quantifying the generation of secondary messengers. However, since the advent of functional selectivity, as well as an increased awareness of several other G protein-independent pathways and the limitations associated with G-protein dependent assays, there is a greater push towards the creation of alternative GPCR ligand screening assays. Towards this endeavor, we describe the application of one such resource, the PRESTO-Tango platform, a luciferase reporter-based system that enables the parallel and simultaneous interrogation of the human GPCR-ome, a feat which was previously considered technically and economically unfeasible. Based on a G-protein independent β-arrestin2 recruitment assay, the universality of β-arrestin2-mediated trafficking and signaling at GPCRs makes PRESTO-TANGO an apt tool for studying approximately 300 non-olfactory human GPCRs, including approximately 100 orphan receptors. PRESTO-Tango's sensitivity and robustness make it suitable for primary high-throughput screens using compound libraries, employed to uncover new GPCR targets for known drugs or to discover new ligands for orphan receptors.
Wprowadzenie
G-protein-coupled receptors (GPCRs) constitute the largest and most diverse family of transmembrane proteins, operating as communication interfaces between a cell and its environment1. The versatility of GPCRs is highlighted by their ability to detect a diverse array of ligands–from neurotransmitters to nucleotides, peptides to photons, and many more–as well as their ability to regulate numerous downstream signaling cascades involved in cellular growth, migration, differentiation, apoptosis, cell firing, etc.2,3. Considering their ubiquity and involvement in a multitude of physiological processes, this receptor family is of utmost therapeutic importance, showcased by the fact that more than a third of currently available prescribed medications target GPCRs4. However, these existing therapeutics only target a small subset of the superfamily (an estimated 10%), and the pharmacology of many GPCRs remains unelucidated. Moreover, more than 100 GPCRs exist as orphan receptors, as they have not been matched with an endogenous ligand5. Thus, GPCR ligand screening is critical in deorphanization and drug development, as it paves the path towards lead discovery and optimization, and possibly to the clinical trial phase.
Methods for GPCR ligand screening have traditionally fallen in one of two categories, G-protein dependent or G-protein independent functional assays6. GPCR signaling is regulated by heterotrimeric G-proteins (Gαβγ), which are activated by the exchange of GTP for GDP bound on the Gα subunit7. Signals from the activated receptor are transduced by G-proteins via secondary messengers, such as cAMP, Calcium, DAG, and IP3, to mediate downstream signaling at downstream effectors8. The nature of the functional consequences of G-protein signaling has been exploited to create cell-based assays that reflect receptor activation. These methods, which measure proximal (direct) or distal (indirect) events in G-protein signaling, are most frequently used for GPCR ligand screening and have been principally employed in deorphanization studies6. Examples of assays that directly measure GPCR-mediated G-protein activation include the [35S]GTPγS binding assay, which measures binding of a radiolabeled and non-hydrolyzable GTP analog to the Gα subunit, and Förster/bioluminescence resonance energy transfer (FRET/BRET, respectively) probes to monitor GPCR-Gα and Gα/Gγ interactions, which have been gaining more traction over the years9,10. Assays that monitor distal events are the most commonly used tools for GPCR profiling; for example, cAMP and IP1/3 assays measure intracellular accumulation of G-protein dependent secondary messengers, whereas [Ca2+] flux and reporter assays involving specific response elements implicated in G-protein activation (CRE, NFAT-RE, SRE, SRF-RE) examine events further downstream the signaling cascade11. While most of the aforementioned assays can be performed at a high-throughput level, are fairly sensitive, and boast certain assay-specific advantages (e.g., discrimination between full/partial agonists, neutral antagonists and inverse agonists in the case of GTPγS binding, or assay functionality on live cells such as [Ca2+] and IP1/3)6, there are unfortunately no existing G-protein dependent methods befitting the interrogation of the entire druggable GPCR-ome. This is largely due to the native coupling of multiple G-protein subfamilies to GPCRs, resulting in signaling at several cascades and the unknown G-protein coupling at orphan GPCRs. To mitigate this issue, assays have been developed to force promiscuous G-protein coupling through a single common signaling read-out, such as cAMP, and Ca2+, albeit most of them are low-throughput12.
An important aspect of the GPCR lifecycle is the termination of G-protein-dependent signaling, which occurs in large part through the recruitment of β-arrestins which induces dissociation of the G-protein, and ultimately desensitizing the receptor, which is targeted for clathrin-coated internalization13. The most ubiquitously expressed isoforms of β-arrestin are the non-visual β-arrestin1 and β-arrestin2, also denoted as arrestin-2 and arrestin-3, respectively14. Enter G-protein independent cell-based assays, which add a new dimension to GPCR ligand screening; receptor trafficking, label-free whole cell, and β-arrestin recruitment assays are all notable examples. GPCR trafficking assays employ fluorophore-labeled ligands or co-internalized antibodies targeting the receptor15, whereas label-free whole cell assays use biosensors which translate cellular changes induced by ligand binding into quantifiable outputs, such as electrical or optical signals16. Notably, quintessential GPCR- β-arrestin interactions fashion the β-arrestin recruitment assay as an attractive tool in the repertoire of functional assays17. The Tango system, first developed by Barnea et al. only a decade ago, involves the introduction of three exogenous genetic elements: a protein fusion consisting of β-arrestin2 with a tobacco etch virus protease (TEVp), a tetracycline transactivator (tTA) that is tethered to a GPCR via a tobacco etch virus protease cleavage site (TEVcs) and is preceded by a sequence from the C-terminus of the V2 vasopressin receptor (V2 tail) to promote arrestin recruitment, and a reporter luciferase gene whose transcription is triggered by the tTA transcription factor translocation to the nucleus, which is freed from the membrane anchoring following β-arrestin2 recruitment (Figure 1)18. Quantitative readings of GPCR activation and β-arrestin2 recruitment can be subsequently determined by reading for luminescence. A notable distinction is that while receptor trafficking and label-free whole cell methods are relatively low-throughput, the Tango has several advantages, including selective read-out that is specific to the target receptor and sensitivity due to signal integration, which make it a suitable candidate for ligand screening on a larger scale18.
In view of these strategic features, Kroeze et al. developed PRESTO-Tango (Parallel Receptor-ome Expression and Screening via Transcriptional Output-Tango), a high-throughput open-source platform that uses the Tango approach to profile the druggable GPCR-ome in a parallel and simultaneous manner19. Exploiting the "promiscuous" recruitment of β-arrestin2 to nearly all GPCRs, PRESTO-Tango is the first-of-its-kind in terms of cell-based functional assays, enabling rapid "first-round" screening of small molecule compounds at almost all non-olfactory GPCRs, including orphans, independent of the G-protein subfamily coupling.
Protokół
1. Primary screening: cell culture and plate seeding
- To prepare poly-L-lysine (PLL)-coated plates, dispense 20 μL/well of a 25 μg/mL stock solution of PLL in white or black 384-well optical bottom plates using an electronic multichannel pipette or a reagent dispenser. Incubate the plates at room temperature for 0.5–2 h.
NOTE: If using the black 384-well plates, expect the background signal to be lower compared to the white plates. Black plates are recommended to reduce bleed-through of luminescence between adjacent wells. - To preserve the coated plates and wash off the excess PLL, remove the PLL by flicking it over the sink, tap dry over a paper towel, and add 40 μL/well of diluted 1x solution of antibiotic-antimycotic using an electronic multichannel pipette or a reagent dispenser. Store PLL-coated plates at 4 °C until ready for plate seeding.
- Maintain HTLA cells (kindly provided by Dr. Richard Axel)–a human embryonic kidney cell line (HEK293T) stably expressing β-arrestin2-TEV and tTA-driven luciferase–in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% of Fetal Bovine Serum, 5% of Bovine Calf Serum, 2.5 μg/mL of puromycin, 50 μg/mL of hygromycin, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a humidified incubator containing 5% CO2.
- Culture HTLA cells in 150 mm dishes and pass cells twice a week at a dilution factor of 1:10, with optimal cell passage number of 5–25. Ensure that a sufficient number of 150 mm dishes are confluent the day of 384-well plate seeding, depending on the scale of the primary screen.
NOTE: Usage of HTLA cells greater than passage 25 may result in reduced viability, yielding suboptimal results. - To seed HTLA cells for the primary screen, gently rinse the confluent 150 mm dish(es) with 1x phosphate-buffered saline (PBS), pH 7.4. Detach cells with approximately 6 mL of 0.05% Trypsin/0.53 mM EDTA, and transfer to a centrifuge tube containing at least equal amount of complete Dulbecco's modified Eagle medium (DMEM) to neutralize the trypsin.
- Spin down HTLA cells at 500 x g for 3 min and resuspend the cell pellet at a density of 0.22 x 106 cells/mL in complete DMEM, omitting the addition of 2.5 μg/mL of puromycin and 50 μg/mL of hygromycin as they can decrease transfection efficacy.
- Incubate the necessary 384-well PLL-coated plates at 37 °C to warm them before seeding cells. Remove the storage solution of 1x antibiotic-antimycotic from the 384-well PLL-coated plate(s) by flicking the plate over the sink and taping it over a paper towel to dry.
- Seed cells into the 384-well PLL-coated plates at a final density of 10,000 cells/well by dispensing 45 μL of the 0.22 x 106 cells/mL HTLA suspension using an electronic multichannel pipet. Incubate plates at 37 °C overnight. If a same-day transfection is preferred, seed cells at a density of 16,000 cells/well and perform the transfection 4 h later.
NOTE: For high transfection efficiency, 50–70% cell confluency is optimal.
2. Primary screening: DNA plate preparation and transfections
- To prepare the 384-well DNA source plate for transfection as shown in Figure 2, distribute the plasmid cDNAs encoding the GPCR-Tango constructs of interest in a 96-well plate, with a different GPCR/well. The plasmid DNA should be suspended in 0.1x Tris-EDTA (TE) buffer at a concentration of 50 ng/μL.
NOTE: The 96-well DNA plates can be sealed and stored at -20 °C, and re-used for multiple screening experiments. All cDNA encoding GPCR-Tango constructs are available commercially (see the Table of Materials) and are cloned in the pcDNA3.1 neomycin plasmid. The PRESTO-Tango GPCR Kit consists of four 96-well plates, which include 80 GPCRs each, a couple of wells with an empty vector as negative controls, and positive control wells that hold the dopamine receptor D2 (DRD2), and wells that carry a plasmid encoding a fluorescent protein (YFP) to track transfection efficiency. - Using a multichannel pipette, manually transfer DNA solution from the 96-well to the a 384-well DNA source plate, adding 10 μL per 384-well. To ensure that each condition of the experiment is assayed in quadruplicate, half of the 96-well DNA plate (rows A-D or E-H) will cover a full 384-well plate by distributing each GPCR in two quadrants (first quadrant = - compound, second quadrant = + compound), such that the same GPCR will be transfected in 8 wells of the 384-well plate (see Figure 2 as a guide).
- Assemble the following transfection reagents needed for calcium phosphate precipitation method, as described by Jordan et al.20: 0.1x TE buffer (1 mM Tris-HCl and 0.1 mM EDTA); 2.5 M CaCl2 solution; 2x Hepes buffer, pH 7.05 (50 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4). Sterilize all the solutions by filtration and store at 4 °C. The day of transfection, allow the reagents to reach room temperature before use.
- Dilute the 2.5 M CaCl2 stock solution in 0.1x TE (1:8 dilution) to a final concentration of 0.313 M CaCl2 and vortex. Transfer 40 μL of 0.313 M CaCl2 to the 384-well DNA source plate and mix by pipetting up and down with a hand-held multichannel pipette or an automated benchtop 384-channel pipettor.
- Add 50 μL of 2x Hepes buffer to the 384-well DNA source plate, mix again by pipetting up and down and let stand for 1 min; each 384-well will have an adequate amount of DNA/transfection mixture for the transfection of nine 384-well plates, depending on the number of compounds that need to be tested. Transfer 10 μL of the DNA/transfection mixture from the 384-well DNA source plate to the seeded HTLA cells and incubate the plates overnight at 37 °C.
3. Primary screening: Cell stimulation
- Twenty-four hours later, decant the transfected cell media by gently flicking the 384-well plate over the sink and taping it over a paper towel, or with an aspirator head. Slowly add 40 μL of starving media (DMEM supplemented with 1% dialyzed fetal bovine serum (dFBS) and 1x antibiotic/antimycotic), being careful to avoid touching the cells directly.
- Pipet 20 μL of the compound of interest at a 3x concentration (final concentration of the drug in the cell plate will be 1x) into the alternating rows with (+) stimulation, and 20 μL of vehicle buffer for the alternating rows without (-) compound. Return the cell plate at 37 °C in 5% CO2 and incubate for at least 16 h.
4. Primary screening: Luminescence reading
- Prepare the Glo reagent, modified from Baker and Boyce21: 108 mM Tris–HCl; 42 mM Tris-Base, 75 mM NaCl, 3 mM MgCl2, 5 mM Dithiothrei-tol (DTT), 0.2 mM Coenzyme A, 0.14 mg/ml D-Luciferin, 1.1 mM ATP, 0.25% v/v Triton X-100, 2 mM Sodium hydrosulfite.
NOTE: Stock solutions of the reagents can be made in advance, except for D-Luciferin, which is always freshly added to the Glo reagent in its powdered form. If black plates were used, the amount of D-Luciferin can be increased up to 0.25 mg/mL. - At 16–24 h following stimulation, decant the transfected cell media by gently flicking the 384-well plate over the sink and taping it over a paper towel. Add 20 μL/well of Glo reagent and incubate the plate at room temperature for 5–20 min. Read the plates using a microplate luminescence counter, with an integration time of 1 s/well.
5. Primary screening: Data analysis
- Export the saved files from the luminescence counter as a spreadsheet; results will be recorded in relative luminescence units (RLU). Based on the layout of the 384-well plate, calculate the activation (fold change) of each receptor using the following formula:
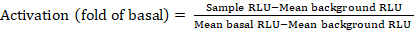
NOTE: Here Sample RLU refers to value of each of the four replicate wells of the stimulated (+ compound) quadrant, Mean background RLU is the mean of the negative controls on the plate, and Mean basal RLU refers to the mean of the untreated quadrant of that same receptor (- compound). In addition, calculate the standard deviation of the 4 data points to verify the quality of the results. It is recommended to perform a log2 transformation on the mean of the fold changes to rectify any heteroskedasticity; the log2 base is a practical choice to help identify positive hits. Empirically set the positive hit thresholds; it has to be noted that some receptors can have as low as 2-fold increase and up to 40-fold increase for others with full agonist. - Based on the results, select the GPCRs that are potential positive hits for secondary screening.
6. Secondary screening: Cell seeding and transfections
- Subculture HTLA cells in 100 mm dishes at a total cell density of 5 x 106 cells in 11 mL of complete media (4.55 x 105/mL) and incubate at 37 °C for 24 h. If a same-day transfection is preferred, seed cells at a density of 7.5 x 106 cells and perform the transfection 4 h later.
- Pre-warm the reagents needed for calcium phosphate precipitation at room temperature. Combine 450 μL of 0.1x TE buffer with 50 μL of 2.5 M CaCl2 and quickly vortex; these amounts are specific for one 100 mm dish, based on the volume of growth medium it holds.
- In a tube, add 500 μL of the TE/CaCl2 solution to 10 μg of GPCR cDNA and vortex. Add 500 µL of 2x Hepes buffer solution in the tube, shake vigorously (do not vortex), and incubate for 1 min.
NOTE: 1 µg of any plasmid encoding a fluorescent protein (e.g. YFP, mCherry, etc.) can be co-transfect with 9 µg of GPCR cDNA for a total of 10 μg. The fluorescent protein is used to track transfection efficiency, and this minimal amount will not interfere with the assay. - Immediately following the short incubation, dispense the 1 mL solution dropwise onto the cells. Gently rock the plate back and forth to evenly distribute the precipitate, taking care not to swirl the plate, and incubate at 37 °C for 24 h.
- The following day, observe the transfection efficiency by looking at the expression of the fluorescent protein under a fluorescent cell imager; transfections greater than 50% coverage are ideal.
- Incubate the necessary 384-well PLL-coated plate(s) in the incubator at 37 °C to warm it before seeding cells. Remove the storage solution of 1x antibiotic-antimycotic from the 384-well PLL-coated plate(s) by flicking the plate over the sink and taping it over a paper towel to dry.
- Gently rinse the transfected cells with Versene solution (1X PBS, pH 7.4; 0.53 mM EDTA), and detach by adding 3 mL of 0.05% trypsin/0.53 mM EDTA to the dish. Transfer the contents to a centrifuge tube containing at least an equal amount of complete DMEM to neutralize the trypsin.
- Spin down the cells at 500 x g for 3 min and resuspend the cells at a density of 0.4 x 106 cells/mL in starving media. Seed cells into the 384-well PLL-coated plate(s) at a final density of 25,000 cells/well by dispensing 45 μL of the cell suspension using an electronic multichannel pipet. Return the plates to the 37 °C for a minimum of 4 h, allowing the cells to properly attach to the wells before proceeding to the stimulation.
7. Secondary screening: Drug plate preparation for 16-point (half log) dose-curve
- In a 96-well plate, add 270 μL of 1X HBSS drug buffer (1x Hank's Balanced Salt Solution [HBSS], 20 mM HEPES pH 7.4, 1x antibiotic-antimycotic), excluding the last row (row H) of the plate, as shown in Figure 4.
NOTE: For peptides, colloidal molecules, and poorly water-soluble compounds, the addition of 0.1–1% BSA is suggested. To prevent drug oxidation, up to 0.01% ascorbic acid can also be added. - From the drug stock, prepare a drug solution (referred to as the "High" concentration) by calculating a final 3x concentration (final concentration of the drug in the cell plate will be 1x). As an example, for a dose–response curve with 10 μM as its highest concentration, prepare the "High" concentration at 30 μM. Pipet 300 μL of "High" concentration into wells in row H.
- In another tube, prepare the "Low" concentration, which represents the "High" concentration divided by 3.16 (half-log). Based on the previous example, the "Low" concentration would be 9.49 μM. Pipet 300 μL of "Low" concentration into wells in row H, adjacent to the "High" wells.
NOTE: The total number of 96-wells needed in row H will depend on the number of cells and stimulation conditions. Four wells (two "High" and two "Low") will have ample drug solution to stimulate an entire 384 well plate. - Perform a serial dilution by pipetting 30 μL of drug solution from the "High" and "Low" wells of row H to the previous row (row G) and mix by manually pipetting up and down, or as recommended, using an electronic multichannel pipette with the "Pipette and Mix" function. Repeat this step up until the first and most diluted row (row A), while discarding tips between dilutions.
NOTE: If desired, the serial dilutions can be stopped before row A, representing an internal control with no drug, in other words, a "true zero". - Using Figure 4 as a reference, stimulate transfected cells by pipetting 20 μL of the "Low" column dilutions from the 96-well plate to rows A–O of the previously seeded 384-well plate, as well as 20 μL of the "High" column dilutions to wells B–P. Incubate the plate at 37 °C for a minimum of 16 h.
8. Secondary screening: Luminescence reading and data analysis
- At 16–24 h following stimulation, decant the transfected cell media by gently flicking the 384-well plate over the sink and taping it over a paper towel. Add 20 μL/well of Glo reagent and incubate the plate at room temperature for 5–20 min. Read the plates using a microplate luminescence counter, with an integration time of 1 s/well.
- Export the saved files from the luminescence counter as a spreadsheet; results will be recorded in relative luminescence units (RLU). Transfer the data of the 384-well plate to a statistics software to analyze the results using its built-in XY analysis for non-linear regression curve fit. Select the built-in 3-parameter dose-response stimulation function "Log(agonist) vs. response (three parameters)",

NOTE: Here Top and Bottom are plateaus in the units of the Y axis, respectively the maximal response and basal level, EC50 is the concentration of the agonist that that generates 50% response between Top and Bottom, and X refers to the log concentration of the agonist. This model assumes that the dose-response curve has a standard Hill slope of 1.
Wyniki
Using the PRESTO-Tango protocol presented herein, a chromaffin granule (CG) extract was screened against 168 non-olfactory GPCR targets, with the majority being orphan receptors. Profiling of said extract was performed by examining β-arrestin2 mobilization at the chosen receptors, based on the principle designed by Barnea et al.18 (Figure 1). Plasmid cDNA of the GPCRs of interest was taken from the PRESTO-Tango GPCR Kit and assembled in two 96 well-plates in the ...
Dyskusje
The conformationally dynamic GPCRs are powerhouses of signal transduction. The physiochemical properties of the binding pockets of these heptahelical receptors, as well as their physiological relevance underscore the need for GPCR ligand screening tools. As presented above, the PRESTO-Tango assay is rapid, sensitive and user-friendly, lending itself to drug development. Not only does this assay measure agonist-induced activation, but it can also be used to quantify the activity of antagonists and allosteric modulators
Ujawnienia
The authours declare no competing interests.
Podziękowania
This work was supported by the Canadian Institutes of Health Research (CIHR grant #MOP142219).
Materiały
| Name | Company | Catalog Number | Comments |
| 384 Well Optical Bottom Plates, Polystyrene Polymer Base, Cell Culture Treated, BLACK, with lid, Sterile | NUNC | 12-566 | |
| 384 Well Optical Bottom Plates, Polystyrene Polymer Base, Cell Culture Treated, White, with lid, Sterile | NUNC | 12-566-1 | |
| 384 Well Round Bottom, Polypropylene, Non-Treated, Blue, non-sterile, without lid | ThermoFisher | 12-565-390 | |
| Antibiotic-Antimycotic | Wisent | 450-115-EL | |
| D-Luciferin, sodium salt | GoldBio | LUCNA | |
| DMEM with L-Glutamine, 4.5g/L Glucose and Sodium Pyruvate | Corning | 10-013-CV | |
| Eppendorf Xplorer, 12-channel, variable, 15–300 µL | Eppendorf | 4861000155 | |
| Eppendorf Xplorer, 12-channel, variable, 5–100 µL | Eppendorf | 4861000139 | |
| Matrix Platemate 2x3 | ThermoFisher | 801-10001 | |
| MicroBeta 1450 Wallac | Perkin Elmer | ||
| Penicilin-Streptomycin | Wisent | 450-201-EL | |
| Poly-L-Lysine hydrobromide | Millipore-Sigma | P2636-500MG | |
| Roth Lab PRESTO-Tango GPCR Kit | Addgene | Kit #1000000068 |
Odniesienia
- Liapakis, G., et al. The G-protein coupled receptor family: actors with many faces. Current pharmaceutical design. 18 (2), 175-185 (2012).
- Pierce, K. L., Premont, R. T., Lefkowitz, R. J. Seven-transmembrane receptors. Nature Reviews Molecular Cell Biology. 3 (9), 639-650 (2002).
- Kroeze, W. K., Sheffler, D. J., Roth, B. L. G-protein-coupled receptors at a glance. Journal of cell science. 116, 4867-4869 (2003).
- Rask-Andersen, M., Almén, M. S., Schiöth, H. B. Trends in the exploitation of novel drug targets. Nature Reviews Drug Discovery. 10 (8), 579-590 (2011).
- Ngo, T., et al. Identifying ligands at orphan GPCRs: current status using structure-based approaches. British journal of pharmacology. 173 (20), 2934-2951 (2016).
- Zhang, R., Xie, X. Tools for GPCR drug discovery. Acta pharmacologica Sinica. 33 (3), 372-384 (2012).
- Kimple, A. J., Bosch, D. E., Giguere, P. M., Siderovski, D. P. Regulators of G-Protein Signaling and Their G Substrates: Promises and Challenges in Their Use as Drug Discovery Targets. Pharmacological Reviews. 63 (3), 728-749 (2011).
- Wettschureck, N., Offermanns, S. Mammalian G Proteins and Their Cell Type Specific Functions. Physiological Reviews. 85 (4), 1159-1204 (2005).
- Denis, C., Saulière, A., Galandrin, S., Sénard, J. -. M., Galés, C. Probing heterotrimeric G protein activation: applications to biased ligands. Current Pharmaceutical Design. 18 (2), 128-144 (2012).
- Yin, H., et al. Lipid G protein-coupled receptor ligand identification using beta-arrestin PathHunter assay. The Journal of Biological Chemistry. 284 (18), 12328-12338 (2009).
- Cheng, Z., et al. Luciferase Reporter Assay System for Deciphering GPCR Pathways. Current Chemical Genomics. 4, 84-91 (2010).
- Roth, B. L., Kroeze, W. K. Integrated Approaches for Genome-wide Interrogation of the Druggable Non-olfactory G Protein-coupled Receptor Superfamily. The Journal of Biological Chemistry. 290 (32), 19471-19477 (2015).
- Jean-Charles, P. -. Y., Kaur, S., Shenoy, S. K. G Protein-Coupled Receptor Signaling Through β-Arrestin-Dependent Mechanisms. Journal of Cardiovascular Pharmacology. 70 (3), 142-158 (2017).
- Smith, J. S., Rajagopal, S. The β-Arrestins: Multifunctional Regulators of G Protein-coupled Receptors. The Journal of Biological Chemistry. 291 (17), 8969-8977 (2016).
- Böhme, I., Beck-Sickinger, A. G. Illuminating the life of GPCRs. Cell Communication and Signaling. 7 (1), 16 (2009).
- Fang, Y. Label-Free Receptor Assays. Drug discovery today. Technologies. 7 (1), 5-11 (2011).
- Wang, T., et al. Measurement of β-Arrestin Recruitment for GPCR Targets. Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences. , (2004).
- Barnea, G., et al. The genetic design of signaling cascades to record receptor activation. Proceedings of the National Academy of Sciences of the United States of America. 105 (1), 64-69 (2008).
- Kroeze, W. K., et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nature Structural & Molecular Biology. 22 (5), 362-369 (2015).
- Jordan, M., Schallhorn, A., Wurm, F. M. Transfecting Mammalian Cells: Optimization of Critical Parameters Affecting Calcium-Phosphate Precipitate Formation. Nucleic Acids Research. 24 (4), 596-601 (1996).
- Baker, J. M., Boyce, F. M. High-throughput functional screening using a homemade dual-glow luciferase assay. Journal of Visualized Experiments. (88), 50282 (2014).
- Li, H., Eishingdrelo, A., Kongsamut, S., Eishingdrelo, H. G-protein-coupled receptors mediate 14-3-3 signal transduction. Signal Transduction and Targeted Therapy. 1 (1), 16018 (2016).
- Walther, C., Ferguson, S. S. G. Minireview: Role of intracellular scaffolding proteins in the regulation of endocrine G protein-coupled receptor signaling. Molecular Endocrinology. 29 (6), 814-830 (2015).
- Gurevich, E. V., Benovic, J. L., Gurevich, V. V. Arrestin2 expression selectively increases during neural differentiation. Journal of Neurochemistry. 91 (6), 1404-1416 (2004).
- Shaikh, S., Nicholson, L. F. B. Optimization of the Tet-On system for inducible expression of RAGE. Journal of Biomolecular Techniques JBT. 17 (4), 283-292 (2006).
- Blau, H. M., Rossi, F. M. Tet B or not tet B: advances in tetracycline-inducible gene expression. Proceedings of the National Academy of Sciences of the United States of America. 96 (3), 797-799 (1999).
- Roche, O., et al. Development of a Virtual Screening Method for Identification of "Frequent Hitters" in Compound Libraries. Journal of Medicinal Chemistry. 45 (1), 137-142 (2002).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone