A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
High-resolution Structural Magnetic Resonance Imaging of the Human Subcortex In Vivo and Postmortem
In This Article
Summary
Here we present a protocol to determine the minimum number images that needed to be registered and averaged to resolve subcortical structures and test whether the individual layers of the LGN could be resolved in the absence of physiological noise.
Abstract
The focus of this study was to test the resolution limits of structural MRI of a postmortem brain compared to living human brains. The resolution of structural MRI in vivo is ultimately limited by physiological noise, including pulsation, respiration and head movement. Although imaging hardware continues to improve, it is still difficult to resolve structures on the millimeter scale. For example, the primary visual sensory pathways synapse at the lateral geniculate nucleus (LGN), a visual relay and control nucleus in the thalamus that normally is organized into six interleaved monocular layers. Neuroimaging studies have not been able to reliably distinguish these layers due their small size that are less than 1 mm thick.
The resolving limit of structural MRI, in a postmortem brain was tested using multiple images averaged over a long duration (~24 h). The purpose was to test whether it was possible to resolve the individual layers of the LGN in the absence of physiological noise. A proton density (PD)1 weighted pulse sequence was used with varying resolution and other parameters to determine the minimum number of images necessary to be registered and averaged to reliably distinguish the LGN and other subcortical regions. The results were also compared to images acquired in living human brains. In vivo subjects were scanned in order to determine the additional effects of physiological noise on the minimum number of PD scans needed to differentiate subcortical structures, useful in clinical applications.
Introduction
The purpose of this research was to test the resolution limits of structural MRI in the absence of physiological noise. Proton density (PD) weighted images were acquired in a postmortem brain over a long duration (two ~24 hr sessions) to determine the minimum number of images that needed to be registered and averaged to resolve the subcortical structures. For comparison, PD weighted images were also acquired in living humans over a number of sessions. In particular, the objective was to ascertain whether it would be possible in a best-case scenario to resolve all six individual layers of the human LGN, which are approximately 1 mm thick (Figure 1).
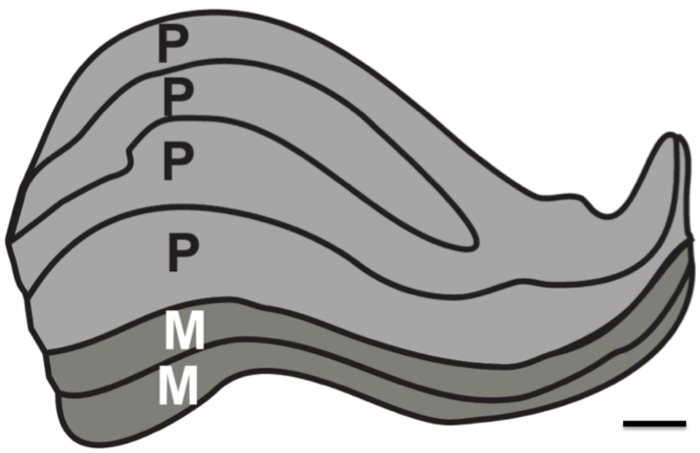
Figure 1. Human Lateral Geniculate Nucleus layers. Schematic of the laminar structure of the LGN. Magnocellular (M) layers are comprised of larger neuronal cell size and smaller cell density that are responsible for resolving motion and course outlines (layers 1-2, depicted as dark grey). Parvocellular layers (P) are comprised of smaller neuronal cell size and larger cell density that are responsible for resolving fine-form and colour (layers 4-6, depicted as light grey). Scale bar 1 mm. Figure based on stained human LGN 12.
Spatial resolution in MRI is improved when the matrix size is increased, and when field-of view (FOV) and slice thickness are decreased. However, increased resolution decreases the signal to noise ratio (SNR), which is proportional to the voxel volume. SNR is also proportional to the square root of the number of measurements. In living humans, although multiple images can be acquired over a number of separate imaging sessions, the ultimate resolution is limited by physiological noise, such as respiration, circulatory pulsations and head movement.
High-resolution (0.35 mm in-plane voxels) PD weighted scans were acquired. PD scans enhance grey and white contrast in the thalamus1, and result in images that minimize T1 and T2 effects. Its image is dependent on the density of protons in the form of water and macromolecules such as proteins and fat in the imaging volume. The increased numbers of protons in a tissue results in a brighter signal on the image due to the higher longitudinal component of magnetization2.
PD-weighted scans were collected since they provide a higher contrast of subcortical structures with surrounding tissue. Other contrasts, such as T1- and T2-weighted images result in difficulty in delineating subcortical structures like the LGN due to smaller contrast-to-noise ratios, as determined ƒ 1,3.
Likewise, earlier studies found that PD-weighted images of formalin fixed post-mortem brains resulted in higher contrast differences between gray and white matter as compared to T1- and T2-weighted images that had similar grey and white matter image intensities 3,4. The underlying biophysical determinants can explain these differences. T1 (longitudinal) and T2 (transverse) relaxation times of hydrogen protons depend on how water moves within the tissue. Fixatives such as formalin work by cross-linking proteins. The differences between water mobility are reduced between different tissue types when fixatives are used. Reduced T1 tissue contrast has been observed after fixation, whereas the differences in the relative density of protons within brain tissues increased with fixation, providing better contrast differentiation 3, 4.
Previous studies have identified the LGN in PD-weighted scans using a 1.5 T 5,6,7, and at 3 T scanner 8,9. It is critical to obtain these scans to be able to precisely outline the extent of the LGN. To maintain full coverage of the subcortical nuclei, 18 PD-weighted slices were obtained within the thalamus. Each volume was re-sampled to twice the resolution 1024 matrix, (0.15 mm in-plane voxel size), concatenated, motion corrected and averaged to produce a high-resolution 3D image of subcortical structures. The optimum number of PD images required for the following slice prescription was 5, reducing scan time to less than 15 min in living humans. Only 1 PD image was required to clearly demarcate subcortical regions in the postmortem brain, reducing scan time to less than 3 min (Figure 2 and 3).
A whole formalin-fixed postmortem brain specimen was scanned from a woman who had died of cardiopulmonary arrest at age 82 years. Review of medical records revealed that she had: chronic obstructive pulmonary disease, angina, triple bypass surgery 8 years prior to death, uterine cancer treated with hysterectomy 7 years prior to death, hyperlipidemia, glaucoma, and cataract surgery. The postmortem brain specimen was immersion-fixed in 10% neutral buffered formalin for at least 3 weeks at 4 °C.The postmortem brain was scanned with the same imaging protocol as well as with other parameters over the course of many hours for image quality comparisons. Only the optimized parameters will be described for the protocol.
Protocol
1. Participant and Postmortem Brain Set-Up
NOTE: All images were acquired using a 3 T MRI scanner with a 32-channel head coil and all MRI scanning was performed at RT, approximately 20 °C. All participants were right handed and gave written informed consent. Each participant was in good health with no history of neurological disorders. The experimental protocol was approved and follows the guidelines of York University Human Participants Review Committee.
- Ask each participant fill out and sign a patient consent form that details MRI safety guidelines and the neuro-imaging protocol.
- For each participant, place earplugs in each ear and secure their head with pads to minimize head motion.
- For post-mortem brain imaging, make sure the brain is fixed prior to neuroimaging and is contained within a bag or container that fits within the MRI head-coil. Place the postmortem brain in the head-coil with its z-axis (superior to inferior) aligned with the bore of the scanner. The brainstem (posterior) should face towards the foot of the scanner bed.
- Place vacuum cushion hands around the post-mortem brain for additional support.
2. Localizing and Prescribing the Subcortex
NOTE: The thalamus is a dual lobed structure located near the center of the brain situated between the midbrain and the cerebral cortex. Located within the dorsal thalamus, the human LGN is a small subcortical structure that extends a maximum of ~10 mm.
- To register a new participant, open the MRI imaging software and click on the Patient tab in the upper left hand corner. Then click on Register.
- Fill in the appropriate patient information, and then click on the Exam tab.
- To obtain a localizer scan, click on the Exam Explorer tab to create a new protocol. Observe the set-up window on the screen, click the Routine tab, and enter the following parameters: acquisition time 28 sec, acquisition matrix 160 × 160, 1 slice, 1.6 mm thick isotropic voxel size, FOV = 260 mm, FoV phase = 100%, slice resolution = 69%, phase and slice partial phase Fourier = 6/8, TR = 3.15 ms, TE = 1.37 msec, Flip Angle = 8°.
- Overlay the slice selection box used for acquiring the PD images over the localizer covering the subcortical nuclei within the thalamus as well as surrounding structures (Figure 4).
3. High-resolution Structural Parameters
- Create a new protocol for obtaining high-resolution PD-weighted scans. In the set-up window on the screen, click the Routine tab, and enter the following parameters in the coronal orientation: acquisition time 179 sec, acquisition matrix 512 × 512, 0.3 × 0.3 × 1 mm3 voxel size, TR = 3.25 sec, TE = 32 msec, flip angle = 120°, interleaved slice acquisition, FoV read = 160 mm, FoV phase = 100%, parallel imaging (GRAPPA) with an acceleration factor of 2.
- Use a Turbo Spin Echo sequence, with an Echo Train Length of 5. The first echo at 32 msec is the effective echo for this sequence. Reduce the bandwidth (BW) to the minimum possible, 40 Hz/pixel, to maximize the SNR. To reduce scan duration, choose 18 slices, each 1 mm thick, with an FOV = 160 mm. This slab provides enough coverage of subcortical regions of interest.
NOTE: For reliable identification of subcortical structures, acquire 5 runs with the above parameters. The total scan duration is only ~15 min (Figure 5). Fat-saturation was not employed.
- Use a Turbo Spin Echo sequence, with an Echo Train Length of 5. The first echo at 32 msec is the effective echo for this sequence. Reduce the bandwidth (BW) to the minimum possible, 40 Hz/pixel, to maximize the SNR. To reduce scan duration, choose 18 slices, each 1 mm thick, with an FOV = 160 mm. This slab provides enough coverage of subcortical regions of interest.
- In post-mortem brain imaging, reliable identification of subcortical structures can be observed in just one scan with the total duration of only ~3 min following the same scanning protocol as in 3.1 (Figure 6).
4. Image Analysis
NOTE: To analyze the MRI data, use the freely available FMRIB's Software Library (FSL) package available for download at (https://www.fmrib.ox.ac.uk/fsl/).
- Open a terminal window, and convert the raw DICOM files from the scanner for each PD volume to a NIfTI format with a DICOM to NIfTI converter. A number of which are freely available for download (e.g., https://www.nitrc.org/projects/mricron). In the command line, type dcm2nii followed by the directory of each PD weighted image run.
- In a terminal window obtain the parameters of the original PD scan. Type fslinfo in the command line followed by the PD scan in NIfTI format.
- Create a high-resolution blank image target volume that has twice the resolution and half the voxel size given by the parameters from fslinfo from the original PD scan. The order of data inputs for this command are as follows:
fslcreatehd <xsize> <ysize> <zsize> <tsize> <xvoxsize> <yvoxsize> <zvoxsize> <tr> <xorigin> <yorigin> <zorigin> <datatype> <headername>
NOTE: For example, if the original PD scan with the following parameters as described in 3.1 are collected (i.e., 512 × 512 matrix, 18 slice, 0.3 × 0.3 × 1 mm3 voxel size, TR = 3.25 s), type the following into the command window:
fslcreatehd 1024 1024 36 1 0.15 0.15 0.5 3.25 0 0 0 4 blankhr.nii.gz - Define the transformation using an identity matrix. Type in any text editor program a text file saved as 'identity.mat' that looks like this:
0 0 0
1 0 0
0 1 0
0 0 1 - Use the flirt command to apply the transformation, upsampling each original PD weighted run to double the total resolution from a 512 to a 1024 matrix, and halve the voxel size in each dimension resulting in a resolution of 0.15 × 0.15 × 0.5 mm3. In a terminal window for each PD volume, type the following flirt command changing the original and output names per run:
flirt -interp sinc -in originalPD.nii.gz -ref blankhr.nii.gz -applyxfm -init identity.mat -out highresPD.nii.gz
NOTE: Where originalPD.nii.gz is the source volume, blankhr.nii.gz is the desired output resolution, and highresPD.nii.gz is the name of the output volume. - Move all the high-resolution images to a new folder, and navigate to it in a terminal window.
- For each participant, concatenate all the upsampled PD images into a single 4D file using fslmerge. In a terminal window type:
fslmerge -t concat_highresPD *.nii.gz
NOTE: This creates a 4D file called concat_highresPD.nii.gz. - Motion correct the concatenated file using mcflirt 10 . This tool allows for an automated robust registration for linear (affine) inter and inter-modal brain images. Select a 4-stage correction, which utilizes sinc interpolation (internally) as a further optimization pass for greater accuracy. In a terminal window type:
mcflirt -in concat_highresPD -out mcf_concat_highresPD.nii.gz -stages 4 -plots
NOTE: This creates a 4D file called mcf_concat_highresPD.nii.gz. - Finally, create the 3D mean image using fslmaths. In a terminal window type:
fslmaths mcf_concat_highresPD.nii.gz -Tmean mean_highresPD.nii.gz
NOTE: This creates a 3D file called mean_highresPD.nii.gz that is of high quality - Visualize the final outcome 3D high-resolution image using the fslview command. In the directory of where your image is, type the following in a terminal window:
fslview mean_highresPD.nii.gz." - Inspect intensity profiles of ROIs in question. Create an ROI using fslview (this can be a vertical line drawn across a region of the LGN for example). In fslview load the high-resolution PD image. Click on the tools tab, then click on the single image tab to enlarge the image for drawing ROIs. Then, click the File tab followed by the Create Mask tab. Draw a line in the ROI of interest. Save the ROI by clicking File, then the Save As. Repeat the line masks for multiple areas within the ROI for intensity comparisons and other ROIs in question.
- Use AFNI's 3dmaskdump command to analyze the resulting intensity of the image. In the directory of where the images are, use the following command in a terminal window to extract the image intensities and location (given as result_mask.txt) of your ROI mask:
3dmaskdump -o result_mask.txt -noijk -xyz -mask ROI_linemask.nii.gz PDaverage_image.nii.gz
Results
Once the subcortex is prescribed within the thalamus, PD weighted images are collected within the slice selection box (Figure 4). The SNR improved by increasing the number of averages in both postmortem and in vivo scans. To determine image quality, the SNR from different scan averages was compared by dividing the signal of the mean brain region by the standard deviation in some area outside the brain. The SNR was calculated as SNR = 0.655 * µtissue/σa...
Discussion
This study describes an optimized protocol in acquisition and analysis technique in order to obtain high-resolution PD weighted images of subcortical regions. A number of scanning parameters were tested and modified with the most significant ones pertaining to matrix size, voxel size, and bandwidth to increase the SNR and decrease the number of acquisitions, a critical step in being able to determine high-resolution subcortical structures. In conjunction with finding the optimal parameters within living humans, this rese...
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge the following funding sources, the Natural Sciences and Engineering Research Council of Canada (NSERC), the Dorothy Pitts Research Fund (NG), and the Nicky and Thor Eaton Research Fund. The authors acknowledge Kevin DeSimone, and Aman Goyal and for their knowledge in MRI acquisition and analysis expertise.
Materials
| Name | Company | Catalog Number | Comments |
| Magnetom Trio 3T MRI | Siemens (Erlangen, Germany). | ||
| Vacuum cushion hand | Siemens | Mat No: 4765454 | Manufactured by: Johannes-Stark-Stk. 8 D-92224 Amberg |
References
- Devlin, J. T., et al. Reliable identification of the auditory thalamus using multi-modal structural analyses. NeuroImage. 30 (4), 1112-1120 (2006).
- Fellner, F., et al. True proton density and T2-weighted turbo spin-echo sequences for routine MRI of the brain. Neuroradiology. 36 (8), 591-597 (1994).
- Schumann, C. M., Buonocore, M. H., Amaral, D. G. Magnetic resonance imaging of the post-mortem autistic brain. J Autism Dev Disord. 31 (6), 561-568 (2001).
- Tovi, M., Ericsson, A. Measurements of T1 and T2 over time in formalin-fixed human whole-brain specimens. Acta Radiol. 33 (5), 400-404 (1992).
- Fujita, N., et al. Lateral geniculate nucleus: anatomic and functional identification by use of MR imaging. Am J Neuroradiol. 22 (9), 1719-1726 (2001).
- Bridge, H., Thomas, O., Jbabdi, S., Cowey, A. Changes in connectivity after visual cortical brain damage underlie altered visual function. Brain. 131 (6), 1433-1444 (2008).
- Gupta, N., et al. Atrophy of the lateral geniculate nucleus in human glaucoma detected by magnetic resonance imaging. Br J Opthalmol. 93 (1), 56-60 (2009).
- Dai, H., et al. Assessment of lateral geniculate nucleus atrophy with 3T MR imaging and correlation with clinical stage of glaucoma. Am J Neuroradiol. 32 (7), 1347-1353 (2011).
- McKetton, L., Kelly, K. R., Schneider, K. A. Abnormal lateral geniculate nucleus and optic chiasm in human albinism. J Comp Neurol. 522 (11), 2680-2687 (2014).
- Jenkinson, M., Bannister, P., Brady, M., Smith, S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 17 (2), 825-841 (2002).
- Dietrich, O., Raya, J. G., Reeder, S. B., Reiser, M. F., Schoenberg, S. O. Measurement of signal-to-noise ratios in MR images: influence of multichannel coils, parallel imaging, and reconstruction filters. J Magn Reson Imaging. 26 (2), 375-385 (2007).
- Andrews, T. J., Halpern, S. D., Purves, D. Correlated size variations in human visual cortex, lateral geniculate nucleus, and optic tract. J Neurosci. 17 (8), 2859-2868 (1997).
- Pfefferbaum, A., Sullivan, E. V., Adalsteinsson, E., Garrick, T., Harper, C. Postmortem MR imaging of formalin-fixed human brain. NeuroImage. 21 (4), 1585-1595 (2004).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved