A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Flow Cytometric Analysis of Natural Killer Cell Lytic Activity in Human Whole Blood
In This Article
Summary
This work describes an advanced workflow for the accurate and fast determination of NK (Natural Killer) cell count and NK cell cytotoxicity in human blood samples.
Abstract
NK cell cytotoxicity is a widely used measure to determine the effect of outside intervention on NK cell function. However, the accuracy and reproducibility of this assay can be considered unstable, either because of user's errors or because of the sensitivity of NK cells to experimental manipulation. To eliminate these issues, a workflow that reduces them to a minimum was established and is presented here. To illustrate, we obtained blood samples, at various time points, from runners (n = 4) that were submitted to an intense bout of exercise. First, NK cells are simultaneously identified and isolated through CD56 tagging and magnetic-based sorting, directly from whole blood and from as little as one milliliter. The sorted NK cells are removed of any reagent or capping antibodies. They can be counted in order to establish an accurate NK cell count per milliliter of blood. Secondly, the sorted NK cells (effectors cells or E) can be mixed with 3,3'-Diotadecyloxacarbocyanine Perchlorate (DiO) tagged K562 cells (target cells or T) at an assay-optimal 1:5 T:E ratio, and analyzed using an imaging flow-cytometer that allows for the visualization of each event and the elimination of any false positive or false negatives (such as doublets or effector cells). This workflow can be completed in about 4 h, and allows for very stable results even when working with human samples. When available, research teams can test several experimental interventions in human subjects, and compare measurements across several time points without compromising the data's integrity.
Introduction
Natural killer cells are an essential element of the innate immune system. While they are very regulated, they have the capability to recognize and eliminate abnormal cells through cell-to-cell contact and without prior activation1. As such, they form a quick line of defense against infections. Exercise, especially intense, has been shown to transiently depress the immune system2,3,4,5. NK cells are particularly prone to this effect4,6,7, effectively creating a window of enhanced sensitivity to disease. Hence, the study of interventions before, during or after intense exercise with the goal of reducing its impact on NK cell function is of particular interest for the well-being of athletes post-competitions.
However, the study of such interventions is complicated by numerous factors: 1) NK cells are sparse8, at about 1% of the white blood cell compartment; 2) NK cells are very sensitive to stress and rely on constant exposure to physiological conditions to remain viable and stable during experimentation; and 3) standard assays to determine NK cell cytotoxicity, such as Ficoll gradients9 and release assays10, are unreliable and inconsistent. The inherent variability of human samples only compound these issues. Fresh human samples collected during interventions are fairly regulated and difficult to procure, at least when compared to animal samples or immortalized cell lines. This reduces opportunities to repeat experiments or add participants to the study cohort to reach significant statistical thresholds. Collectively, these issues support the need for a streamlined protocol that allows for both high-throughput and a high-reliability analysis of NK cell lytic activity in human samples.
We established a workflow that shortens the time necessary to identify, isolate and test NK cells from human whole blood while minimizing exposure to extraneous factors. The method optimizes the use of two instruments, a magnetic-based cell sorter and an imaging flow cytometer, and an assay-specific, optimized T:E ratio to allow the detection of decreases or increases of NK cell cytotoxicity.
Protocol
NOTE: All blood collection procedures were conducted in accordance with the guidelines set forth by the Appalachian State University (ASU) Institutional Review Board (IRB).
1. Whole Blood Collection
- Have a certified phlebotomist draw blood according to World Health Organization's guidelines.
- Draw blood into one 4 ml blood collection tube containing Di-Potassium Ethylenediaminetetraacetic acid (K2EDTA). Invert blood collection tube according to the manufacturer's instructions. Keep blood collection tube at room temperature on a bench-top rocker until separation.
2. Preparation of DiO-labeled Target Cells
- Grow K562 cells in Complete Iscove's Modified Dulbecco's Media (IMDM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin streptomycin for several weeks prior to the assay to ensure the health of the cells. Adjust the concentration of the cells to 3 x 105 cells/mL daily through the completion of a cell count and subsequent passage of cells. Perform a complete media change every 2 to 3 days.
- On the day of the assay, perform a cell count using a 1:1 hemocytometer.
- Remove 10 µL from the K562 flask and place into 1.5 mL tube.
- Add 10 µL of trypan blue dye into the same 1.5 ml tube for a dilution factor of 1:1.
- Gently flick tube to mix K562 cells and trypan blue dye.
- Allow K562 cell-trypan blue dye to incubate for 1 min at room temperature.
- Remove 15 µL of stained K562 cells from tube.
- Pipette onto hemocytometer for cell count.
- Count cells in the four corner squares as well as the middle square for a total of five squares.
- Calculate cell count by using the equation:
Total cells/mL = Total cells counted X (dilution factor/Number of squares) X 10,000/mL
- Resuspend K562 target cells at a final density of 1 x 106 cells/ml in serum free IMDM culture medium.
- For unstained K562 target cells, add 10 mL of K562 target cells to one 15 mL tube at a density of 1 x 106 cells/mL for a total of 10 x 106 K562 cells. Place into a 37 °C incubator with 5% CO2 until ready for use.
- For DiO stained K562 target cells, add 10 mL of K562 targets cells to one 15 mL tube at a density of 1 x 106 cells/mL for a total of 10 x 106 K562 cells.
- Add 1 µL of DiO cell-labeling solution per mL of cell suspension in 15 ml tube designated for DiO staining and gently vortex. For example, add 10 µL of DiO cell-labeling solution to 10 x 106 K562 cells/ml at a volume of 1 x 106 cells/mL.
- Incubate K562-DiO solution for 20 min at 37 °C with 5% CO2 in a 15 ml tube.
- Following incubation, add 3 ml of room temperature phosphate-buffered saline (PBS) to K562-DiO solution containing 15 ml tube.
- Centrifuge for 10 min at 135 x g at room temperature.
- Carefully remove supernatant without disturbing the cell pellet with a 1,000 µl volume-adjustable pipette.
- Add 10 mL of fresh IMDM to cell pellet-containing 15 mL tube.
- Gently vortex tube to resuspend the cells.
- Repeat steps 2.7 to 2.10 two more times.
- Store cells in a 37 °C incubator with 5% CO2 until ready for use.
NOTE: Cells can be stored in the incubator for up to 24 h but it is preferable to use them on the same day.
3. Preparation of Controls
- Transfer the following into separate and appropriately labeled 1.5 ml tubes:
- Add 500 µL of fresh IMDM containing resuspended DiO-labeled K562 cells into the "Double Positive" labeled 1.5 mL tube.
- Add 500 µL of fresh IMDM containing resuspended DiO-labeled K562 cells into the "DiO only" labeled 1.5 mL tube.
- Add 500 µL of fresh IMDM containing resuspended unlabeled K562 cells into the "Propidium Iodide (PI) only" labeled 1.5 mL tube.
- Place the Double Positive and PI only tubes in a 55 °C water bath for 5 min.
- After the 5 min has elapsed, remove tubes and wipe down with 70% ethanol.
- Add 10 L of PI to the Double Positive and PI only 1.5 mL tubes.
- Place all three K562 target cell controls in the incubator at 37 °C for 30 min.
- After the 30 min incubation has elapsed, centrifuge all three K562 target cell controls for 2 min at 163 x g.
- Carefully remove supernatant without disturbing the cell pellet.
- Resuspend each control with 20 µL of fresh IMDM cell culture media, and leave in the 37 °C incubator with 5% CO2 for at least 30 min for optimal DiO signal.
NOTE: Controls are now ready to be run through the imaging flow cytometer.
4. NK Cell Automated Separation
- Turn on cell separator and allow the start-up cycle to finish.
- Ensure that all fluid bottles illuminations are green and that the waste bottle is empty.
- Obtain a room temperature 15 mL tube rack.
NOTE: Selection is based on sample volumes. For example, for a volume less than 3 ml use a 15 ml tube rack and for a volume more than 3 mL use a 50 mL tube rack. - Label sample tubes accordingly (Repeat per sample/participant): Participant 1 Whole Blood Sample; Participant 1 Negative fraction; Participant 1 Positive fraction.
- Gently pipette 1.5 mL of whole blood from Step 1.2 into "Whole Blood Sample" 15 mL tube.
- Place appropriately labeled 15 mL "Whole Blood Sample" tube from Step 4.5 and labeled 15 mL "Negative Fraction" and "Positive Fraction" tubes from Step 4.4 in the tube rack. Use thefollowing sample rack set-up: Row (R1) A: Whole Blood Sample, Row (R2) B: Negative, unlabeled fraction, Row (R4) C: Positive, magnetically labeled fraction.
- Insert the separator rack onto the MiniSampler for autolabeling.
- Select "Reagent" on the menu and highlight the position where the vial will be placed in the separator rack.
- Select "Read Reagent" to activate the 2D code reader.
- Place the appropriate reagent vial in front of the 2D code reader between 0.5-2.5 cm from the code reader cover.
NOTE: For example, the required reagent for NK cell separation is CD56. - Hold reagent vial at an angle in front of 2D code reader for optimal reading.
- Place the reagent vial into the proper separator rack position.
- Highlight the desired positions under the Separation tab on the screen.
- From the Labeling submenu, assign an autolabeling program.
- Assign the Cell Separation Reagent CD56 MicroBeads to rack positions 1, 2, 3, and 4.
- Select the "posselwb" separation program.
- Select the "rinse" wash program.
- Insert a sample volume of 1,500 µL in the "Volume" submenu using the numeric keypad.
- Select the "Enter" option on the keypad.
- Select "Run" to start the cell separation.
- Select "OK" to confirm that enough buffer is available for processing all samples.
5. NK Cell Count Following Cell Separation
- Immediately following cell separation with the cell separator, retrieve the positive fraction. Leave at room temperature. This fraction contains the desired pure NK cell population.
- For each individual sample, perform a cell count using a hemocytometer as per Step 2.2.
- After calculations, record the cell count.
6. Cytotoxicity Assay Sample Preparation
- Prepare and label 1.5 mL tubes for each sample/participant accordingly.
- Pipette desired ratio of NK cells and DiO-labled K562 cells into each tube.
NOTE: For example, the desired ratio of K562 target cells and NK effector cells is 1:5. - Centrifuge for 5 min at 135 x g.
- Carefully remove supernatant without disturbing the cell pellet.
- Resuspend NK-DiO labeled K562 cell mixture in 500 µL of NK cell media without interleukin-2 (IL-2) and 2-mercaptoethanol (2-ME) (incomplete NK cell culture media).
NOTE: The incomplete NK cell culture media is Minimum Essential Medium Eagle with sodium bicarbonate, without L-glutamine, ribonucleosides and deoxyribonucleosides. - Add 5 µL of PI to each tube.
- Centrifuge for 2 min at 163 x g.
- Incubate cells at 37 °C for 2 h.
- Following 2 h incubation, centrifuge for 2 min at 163 x g.
- Carefully remove supernatant without disturbing the cell pellet.
- Resuspend cells in 25 µL incomplete NK cell culture media.
7. Preparation of Spontaneous ("S") Sample
- Pipette 500 µl of DiO-labeled K562 cells (concentration of 1 x 106 cells/mL) into 1.5 mL tube.
- Add 10 µL of PI to each tube.
- Centrifuge tube for 2 min at 163 x g.
- Incubate cells at 37 °C for 2 h.
- Following 2 h incubation, centrifuge for 2 min at 163 x g.
- Carefully remove supernatant without disturbing the cell pellet.
- Resuspend cells in 25 µl incomplete alpha-Minimum Essential Medium (α-MEM) cell culture media.
8. Data Acquisition with Imaging Flow Cytometer
- Press the green button inside the front door of the imaging flow cytometer to turn on the instrument.
- Turn on all computers associated with the imaging flow cytometer.
- Launch the imaging flow cytometer software.
- Click the "Startup" button to flush the system and prepare the sample line.
- Once the "Startup" is complete, close out the "Calibrations" window.
- Assign channels: on the top left-hand side, click on each channel in order to assign them.
- On the right hand side, click on the scatter plot button to create 4 scatterplots: Raw Max Pixel _MC_6 vs Area_M06, Raw Max Pixel _MC_2 vs Area_M02, Raw Max Pixel _MC_5 vs Area_M05 and FieldArea_M01 vs AspectRatio_M01.
- Begin analyzing samples by first using the "Double Positive" control.
- Click on "Load."
- Place the 1.5 ml tube with the "Double Positive" sample from Steps 3.4 to 3.9 into the sample loader.
- Select the 40X objective under the "Magnification" tab.
- Turn on the 405 mW and 642 mW lasers.
- Turn on the "Brightfield" channel.
- Click on "Select Intensity."
- Based on the "Double Positive" control sample, determine the desired intensity for the 405 mW laser so the detector is not overloaded.
Note: For example, the optimal intensity for this experiment was set at 11 mW.
- After the desired set-up is achieved, acquire data.
- Under the "File Acquisition" tab, enter in a custom filename text. Select a folder for saving the data file(s).
- Enter the number of cells to acquire next to "Collect". Typically this number varies between 1,000 to 10,000.
- Click on "Acquire."
NOTE: Once the desired number of cells is acquired, the data file is automatically saved in the previously selected folder.
- After acquisition finishes, load the next control sample – DiO only control.
- Click on "Load."
- Place the 1.5 mL tube with the "DiO only" sample into the sample loader.
- Leave the 40X objective under the "Magnification" tab selected.
- Leave the 405 mW laser turned on.
- Turn OFF the 642 mW laser and the "Brightfield" channel.
NOTE: Now that the desired set-up has been achieved, data can be acquired. - Under the "File Acquisition" tab, enter in a custom filename text and select a folder for saving the data file(s). Enter the number of cells to acquire next to "Collect.". Typically this number is 1,000.
- Click on "Acquire."
NOTE: Once the desired number of cells is acquired the data file is automatically saved in the previously selected folder.
- Repeat step 8.10 for the "PI only" control sample. The experimental samples are now ready to be collected.
- Handle the remaining experimental samples, including the "Spontaneous 'S' Samples" as follows:
- Leave the 40X objective under the "Magnification" tab selected.
- Turn ON the 405 mW and 642 mW lasers.
- Turn ON the "Brightfield" channel.
- Click on "Set Intensity."
- Under the "File Acquisition" tab, enter in a custom filename text and select a folder for saving the data file(s). Enter in a custom filename text.
- Enter the number of cells to acquire next to "Collect."
- Click on the "Acquire" button.
NOTE: Once the desired number of cells is acquired the data file is automatically saved in the previously selected folder.
- Repeat step 8.12 for all experimental samples.
- After all experimental data and files have been collected, click the "Shutdown" button to sterilize the system.
9. Imaging Flow Cytometer Sample Analysis
- Open the imaging flow cytometer analysis software application.
- Under "File", open an experimental .RIF file.
- Build a new matrix using the single color .RIF files (DiO-only control and PI-only control, created during steps 8.10 and 8.11) by selecting "Create a New Matrix" under the Compensation tab in the imaging flow cytometer software.
NOTE: The software will prompt for the selection of the single color files and merge them to create a matrix file (.ctm file extension), that is to be selected to apply channel compensation. - Create dot plots by using the "building blocks" function of the software.
- Create a dot-plot (BrightFieldGradient_RMS vs frequency) to gate the focused cells. Call the gate "Focus" (Figure 1A).
- Using the "Focus" gate, create a dot plot of Bright Field Area vs Bright Field Aspect Ratio to gate the singlet cells. Call the gate "Single" (Figure 1B).
- Using the "Single" gate, create a dot plot of Intensity_MC_Ch02 vs Intensity_MC_Ch5. Use this plot to identify and gate DiO-positive only cells (targets, alive) and PI-DiO double positive cells (targets, dead) (Figure 1C).
NOTE: All the plots described in steps 9.4.1, 9.4.2, and 9.4.3 can be created by using the "building blocks" function of the software.
- Click on the statistics function of the dot plot to access cell numbers of each gate.
- Calculate the percentage of dead targets in the spontaneous sample and experimental samples using the following formula:
% dead targets in sample = (#dead targets x 100)/(#live targets + #dead targets) - Calculate cytotoxicity using the following formula:
% cytotoxicity = [(Experimental dead-Spontaneous dead)/(100-Spontaneous dead)]x100
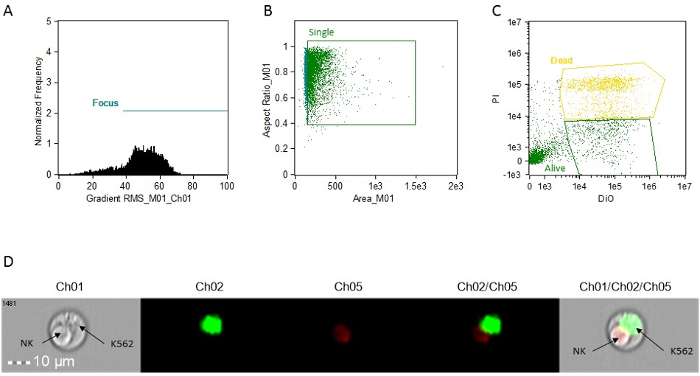
Figure 1: Representative histograms,scatter plots and images for cytotoxic activity analysis. (A) focus cell analysis. (B) single cell analysis. (C) target cell staining analysis. All determinations are made using the image attached to each event. This can be accessed in analysis software by simply clicking on the event on the graphs. (D) representative image of a doublet event, showing an apoptotic NK cell and a live K562 target. Ch01, Brightfield. Ch02, DiO. Ch05, PI. Please click here to view a larger version of this figure.
Results
Determination of NK cell count
The effect of heavy running on NK cell count in whole blood was measured, using the exercise protocol described in Figure 2. Blood samples were drawn before exercise, immediately after exercise, 1.5 h after exercise, and finally 24 and 48 h after the initial blood draw. The concentration of NK cells per milliliter of whole blood was measured for each runner (Figure 3A) and on average (Figure 3B) for ea...
Discussion
The method described in this study directly measures the specific activity of an individual's NK cells in response to stimuli (in this particular case, prolonged exercise). Typically, NK cells are isolated from one's blood using density gradients or cell sorting by using a combination of markers. While these methods are widely used, they have many drawbacks: they are time consuming, involve multiple manipulations, and are heavily user dependent. As a result, undue stress is placed on the isolated NK cells, which ...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This project was supported by Agriculture and Food Research Initiative Competitive Grant no. 2100-68003-30395 from the USDA National Institute of Food and Agriculture.
Materials
| Name | Company | Catalog Number | Comments |
| K-562 lymphoblasts | ATCC | CCL-243 | |
| Iscove's Modified Dulbecco's Media | ATCC | 30-2005 | High glucose, with L-Glutamine, with HEPES, Sterile-filtered |
| Alpha Minimum Essential medium | ATCC | CRL-2407 | Without ribonucleosides and deoxyribonucleosides but with 2 mM L-glutamine and 1.5 g/L sodium bicarbonate |
| Trypan Blue Solution 0.4% | Amresco | K940-100ML | Tissue culture grade |
| Propridium Iodide Staining Solution | BD Pharmingen | 51-66211E | |
| Vybranto DiO cell-labeling solution | Vybranto | V-22886 | |
| autoMACS Pro Separator | Miltenyi Biotec | 130-092-545 | |
| autoMACS Running Buffer | Miltenyi Biotec | 130-091-221 | |
| autoMACS Washing Buffer | Miltenyi Biotec | 130-092-987 | |
| autoMACS Columns | Miltenyi Biotec | 130-021-101 | |
| Whole Blood CD56 MicroBeads, human | Miltenyi Biotec | 130-090-875 | |
| ImageStream X Mark II Imaging Flow Cytometer | EMD Millipore | ||
| Speedbeads | Amnis Corporation | 400030 | |
| 0.4-0.7% Hypochlorite (Sterilizer) | VWR | JT9416-1 | |
| Coulter Clenz | Beckman Coulter | 8546929 | |
| 70% Isopropanol (Debubbler) | EMD Millipore | 1.3704 | |
| D-PBS (Sheath fluid) | EMD Millipore | BSS-1006-B (1X) | No calcium or magnesium |
| INSPIRE Software | EMD Millipore | Version Mark II, September 2013 | |
| Ideas Application Software | EMD Millipore | Version 6.1, July 2014 |
References
- Cerwenka, A., Lanier, L. L. Natural killer cells, viruses and cancer. Nat Rev Immunol. 1 (1), 41-49 (2001).
- Nieman, D. C. Exercise, infection, and immunity. Int J Sports Med. 15, 131-141 (1994).
- Romeo, J., Warnberg, J., Pozo, T., Marcos, A. Physical activity, immunity and infection. Proc Nutr Soc. 69 (3), 390-399 (2010).
- Nieman, D. C. Marathon training and immune function. Sports Med. 37 (4-5), 412-415 (2007).
- Simpson, R. J., Kunz, H., Agha, N., Graff, R. Exercise and the Regulation of Immune Functions. Prog Mol Biol Transl Sci. 135, 355-380 (2015).
- Walsh, N. P., et al. Position statement. Part one: Immune function and exercise. Exerc Immunol Rev. 17, 6-63 (2011).
- Timmons, B. W., Cieslak, T. Human natural killer cell subsets and acute exercise: a brief review. Exerc Immunol Rev. 14, 8-23 (2008).
- Westermann, J., Pabst, R. Distribution of lymphocyte subsets and natural killer cells in the human body. Clin Investig. 70 (7), 539-544 (1992).
- Boyum, A. Isolation of leucocytes from human blood. Further observations. Methylcellulose, dextran, and ficoll as erythrocyteaggregating agents. Scand J Clin Lab Invest Suppl. 97, 31-50 (1968).
- McMillan, R., Scott, J. L. Leukocyte labeling with 51-Chromium. I. Technic and results in normal subjects. Blood. 32 (5), 738-754 (1968).
- Berk, L. S., et al. The effect of long endurance running on natural killer cells in marathoners. Med Sci Sports Exerc. 22 (2), 207-212 (1990).
- McAnulty, L. S., et al. Effect of blueberry ingestion on natural killer cell counts, oxidative stress, and inflammation prior to and after 2.5 h of running. Appl Physiol Nutr Metab. 36 (6), 976-984 (2011).
- Millard, A. L., et al. Brief Exercise Increases Peripheral Blood NK Cell Counts without Immediate Functional Changes, but Impairs their Responses to ex vivo Stimulation. Front Immunol. 4, 125 (2013).
- Janeway, C. A., Travers, P., Walport, M., Shlomchik, M. J. . Immunobiology: The Immune System in Health and Disease. 5th edn. , (2001).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved