Method Article
A Metadata Extraction Approach for Clinical Case Reports to Enable Advanced Understanding of Biomedical Concepts
In This Article
Summary
We present a protocol and associated metadata template for the extraction of text describing biomedical concepts in clinical case reports. The structured text values produced through this protocol can support deep analysis of thousands of clinical narratives.
Abstract
Clinical case reports (CCRs) are a valuable means of sharing observations and insights in medicine. The form of these documents varies, and their content includes descriptions of numerous, novel disease presentations and treatments. Thus far, the text data within CCRs is largely unstructured, requiring significant human and computational effort to render these data useful for in-depth analysis. In this protocol, we describe methods for identifying metadata corresponding to specific biomedical concepts frequently observed within CCRs. We provide a metadata template as a guide for document annotation, recognizing that imposing structure on CCRs may be pursued by combinations of manual and automated effort. The approach presented here is appropriate for organization of concept-related text from a large literature corpus (e.g., thousands of CCRs) but may be easily adapted to facilitate more focused tasks or small sets of reports. The resulting structured text data includes sufficient semantic context to support a variety of subsequent text analysis workflows: meta-analyses to determine how to maximize CCR detail, epidemiological studies of rare diseases, and the development of models of medical language may all be made more realizable and manageable through the use of structured text data.
Introduction
Clinical case reports (CCRs) are a fundamental means of sharing observations and insights in medicine. These serve as a basic mechanism of communication and education for clinicians and medical students. Historically, CCRs have also provided accounts of emerging diseases, their treatments, and their genetic backgrounds1,2,3,4. For example, the first treatment of human rabies by Louis Pasteur in 18855,6 and the first application of penicillin in patients7 were both reported through CCRs. More than 1.87 million CCRs have been published as of April 2018, with over half a million within the last decade; journals are continuing to provide new venues for these reports8. Though unique in form and content, CCRs contain text data that are largely unstructured, contain a vast vocabulary, and concern interrelated phenomena, limiting their use as a structured resource. Significant effort is required to extract detailed metadata (i.e., "data about data", or in this case, descriptions of document contents) from CCRs and establish them as a findable, accessible, interoperable, and reusable (FAIR)9 data resource.
Here, we describe a process for extracting text and numerical values to standardize the description of specific biomedical concepts within published CCRs. This methodology includes a metadata template to guide annotation; see Figure 1 for an overview of this process. Application of the annotation process to a large collection of reports (e.g., several thousand of a specific type of disease presentation) permits assembly of a manageable and structured set of annotated clinical texts, achieving machine-readable documentation and biomedical phenomena embedded within each clinical presentation. Though data formats such as those provided by HL7 (e.g., Version 3 of the Messaging Standard10 or the Fast Healthcare Interoperability Resources [FHIR]11), LOINC12, and revision 10 of the International Statistical Classification of Diseases and Related Health Problems (ICD-10)13 provide standards for describing and exchanging clinical observations, they do not capture the text surrounding these data, nor are they intended to. The results of our methodology are best used to enforce structure on CCRs and facilitate subsequent analysis, normalization through controlled vocabularies and coding systems (e.g., ICD-10), and/or conversion to the clinical data formats listed above.
Mining CCRs is an active area of work within biomedical and clinical informatics. Though previous proposals to standardize the structure of case reports (e.g., using HL7 v2.514 or standardized phenotype terminology15) are commendable, it is likely that CCRs will continue to follow a variety of different natural-language forms and document layouts, as they have for much of the past century. Under ideal conditions, authors of new case reports follow CARE guidelines16 to ensure they are comprehensive. Approaches sensitive to both natural language and its relation to medical concepts may therefore be most effective in working with new and archived reports. Resources such as CRAFT17 and those produced by Informatics for Integrating Biology and the Bedside (i2b2)18 curation support natural language processing (NLP) approaches yet do not specifically focus on CCRs or clinical narratives. Similarly, medical NLP tools such as cTAKES19 and CLAMP20 have been developed but generally identify specific words or phrases (i.e., entities) within documents rather than the general concepts commonly described in CCRs.
We have designed a standardized metadata template for features commonly included within CCRs. This template defines features to impose structure on CCRs—an essential precursor for in-depth comparisons of document contents-yet allows for sufficient flexibility to retain semantic context. Though we have designed the format associated with this template to be appropriate for both manual annotation and computationally-assisted text mining, we have ensured it is particularly easy to use for manual annotators. Our approach noticeably differs from more intricate (and, therefore, less immediately understandable to untrained researchers) frameworks such as FHIR21. The following protocol describes how to isolate document features corresponding to each template data type, with a single set of values corresponding to those in a single CCR.
The data types within the template are those most descriptive for CCRs and patient-focused medical documents in general. Annotation of these features promotes findability, accessibility, interoperability, and reusability of CCR text, primarily by giving it structure. The data types are in four general categories: document and annotation identification, case report identification (i.e., document-level properties), medical content concepts (primarily concept-level properties), and acknowledgements (i.e., features providing evidence of funding). In this annotation process, each document includes the full text of a CCR, omitting any document contents material independent to the case (e.g., experimental protocols). CCRs are generally less than 1,000 words each; a single corpus should ideally be indexed by the same bibliographic database and be in the same written language.
The product of the approach described here, when applied to a CCR corpus, is a structured set of annotated clinical text. While this methodology can be performed fully manually and has been designed to be performed by domain experts without any informatics experience, it complements the natural language processing approaches specified above and provides data appropriate for computational analysis. Such analyses may be of interest to audiences of researchers beyond those who frequently read CCRs, including:
- those concerned with disease presentations, their key symptomology, usual diagnostic approaches, and treatments
- those who wish to compare the results of clinical trials with events described within the clinical literature, potentially providing additional observations and greater statistical power.
- bioinformatics, biomedical informatics, and computer science researchers who require structured medical language data sets or high-level understandings of medical narratives
- Government policy researchers focusing on how clinical trials may best reflect how diagnosis and treatment as it occurs in reality
Enforcing structure on CCRs can support numerous subsequent efforts to better understand both medical language and biomedical phenomena.
Protocol
1. Document and Annotation Identification
Note: Values in this category support the annotation process.
- Using the annotation template, provide an identifier specific to this metadata set, e.g., Case123. The identifier format should be consistent throughout the project (e.g., Case001 through Case500).
- Specify the date on which a document was read and annotated. Use a format resembling “Jan 10 2018” for consistency and readability.
2. Case Report Identification
Note: Values in this category provide document-level features and contribute to a document’s findability.
- Be consistent with the format of each field across all annotations, e.g., individual values should be separated by semicolons without following spaces in all entries. Use identical formats to those used in the original document or those used in a bibliographic database such as MEDLINE.
- Provide the title of the document.
- Provide the names of all authors of the document in the provided order. Normalize the format of all names, such that all names take the form of a single last name followed by any number of initials, e.g. Jane B. Park becomes Park JB. Do not include titles. Separate multiple authors with a semicolon without additional punctuation, such that John A. Smith, Jane B. Park takes a form of Smith JA;Park JB.
- Provide the year of publication of the document.
- Provide the full title of the journal in which the document was published. A list of controlled journal names is provided by the NLM Catalog (https://www.ncbi.nlm.nih.gov/nlmcatalog).
- Provide the address of the home institution of the authors of the document, as specified in the document. This may include departments, geographic locations, and postal address details.
- If multiple locations are provided (e.g., if affiliations differ between authors), specify only details for the corresponding author. If a corresponding author cannot be identified, use that of the first author, or do not specify an institution. If a corresponding author has multiple affiliations, specify both and separate with a semicolon.
- Provide the corresponding author of the document, as specified within the document heading using the same format as that used in the Authors data type.
- Provide a document identifier (e.g., a PMID).
- Provide a Digital Object Identifier, where possible and available, resolvable to the document URL (through https://www.doi.org/), not a PubMed Central page.
- Provide a stable URL to the full text of the document, if available. To maximize accessibility, this may refer to the PubMed Central version.
- Provide the document language. For documents available in multiple languages, provide both, separated with a semicolon.
3. Medical Content
Note: Values in this category identify document-level, concept-level, and text-level features. They serve to enhance a document’s accessibility, interoperability, and reusability. These features provide ways to observe conceptual and semantic similarities between document content, with a focus on biomedical topics and events. Most categories in this section can include multiple text statements and each should be separated using a semicolon.
- Include contextual detail in each field (e.g., “mother had breast cancer at age 50”) rather than providing only terms from a controlled vocabulary (e.g., not “breast cancer” alone). Do not include extensive detail beyond each observation.
- Omit commonly repeated words and phrases (e.g., pronouns, the word “patient”, and the phrases “complained of” or “presented with”). Though subjectivity across multiple annotators is likely, it may be reduced by having multiple annotators for each document and through automated normalization after data collection. Computational post-processing approaches will vary by subsequent analysis needs and are not discussed here in detail.
- Provide the following information in the annotation template.
- Provide specific terms identified within a document, usually in its header, as key terms. Separate with a semicolon as terms may include other punctuation.
- Provide demographic values, specifically any text statements describing a patient's background, including sex and/or gender, age, ethnicity, or nationality.
- Provide geographic locations mentioned within the clinical narrative, other than specific institution addresses. This should not include anatomical locations/parts, but may include any geographic locale where the patient resides or travels.
- Provide life style values, including any text statements describing frequent patient activities or behaviors relevant to their general health. In practice, this frequently involves smoking or alcohol consumption habits, but may also include sun exposure, diet, or frequency of specific types of physical activity.
- Provide medical history values referring to family history. Include any text statements describing clinical observations of and events experienced by siblings, parents, and other family members. This includes genetic conditions and negative observations (i.e., family history was negative for a disease).
- Provide values referring to Social History, including any text statements describing patient background not covered in Demography or Life Style. There may be overlaps in content between these categories. The statements may include occupational history and social habits.
- Provide values referring to the patient’s medical and surgical history. Include any text statements describing any medical observations, treatments, or other events taking place prior to the beginning of the clinical presentation. This includes obstetric history and periods of good health, where noted.
- Specify one or more of the following 16 disease system categories. Note that these values are categorical rather than free-text. Categories are not comprehensive but should indicate most systems impacted by the events described in the clinical presentation and diagnosed disease.
- Follow a specific set of categories, based on the categories used in the International Statistical Classification of Diseases and Related Health Problems, revision 10 (ICD-10) code system. See Table 1 for the list of disease system categories along with corresponding ICD-10 code ranges.
- Provide details of all signs and symptoms. Include any text statements describing any medical observations of signs or symptoms beginning at initial presentation, including their onset, duration, severity, and resolution, if provided. Do not include symptoms described in the outcome. These values may overlap with other types if symptoms continue from history to initial presentation.
- Provide details of any comorbidities. Include any terms or phrases describing distinct diseases present at the time of initial clinical presentation. There is likely overlap between these values and those in clinical history, though Comorbidity should not include terms identical to those in the Diagnosis.
- Provide details of all diagnostic techniques and procedures. Include the names of medical procedures done for diagnostic purposes, including examinations, tests, and imaging, as well as the conditions under which these tests were performed and relevant anatomical locations (e.g., “upper extremity venous ultrasound”). Exclude test results.
- Provide details of diagnosis. Include any text statements describing diagnoses of disease, even if the final diagnosis is ambiguous.
- Provide all laboratory values and test results. Include names of diagnostic tests, their values, and conditions under which they were performed. This will involve overlap with terms used in the Diagnostic Techniques and Procedures data type. Both numerical and qualitative values (e.g., complete blood count was within normal limits) are acceptable. If the names of diagnostic tests are not provided, use terms describing the results (e.g., leukopenia), though they should also be included in the Signs and Symptoms.
- Provide details of pathology. Include any text statements describing results of pathology and histology studies, including gross pathology, immunology, and microscopy studies. Terms may overlap with those used in Diagnostic Techniques and Procedures (step 3.11), e.g., with the procedures performed to obtain samples such as biopsy.
- Provide all pharmacological therapies. Include any text statements describing drug therapies used in the course of treatment, including general terms such as antibiotics or specific drug names. Also, include descriptions of when and how drug therapies were stopped.
- Provide all interventional procedures. Include any text statements describing therapeutic procedures used in the course of treatment, including invasive procedures, implantation of medical devices, and procedures done to facilitate other therapies. Also, include descriptions of when and how ongoing therapeutic procedures were stopped, if necessary.
- Provide the patient outcome. Include any text statements describing health of the patient as of the end of the clinical presentation described in the report, including any follow-up tests.
- Provide counts of all diagnostic images, figures, videos/animations, and tables. Include all counts of visual media included in the report, in the following format: Count of images; Count of figures; Count of videos or animations; Count of tables.
- Distinguish between images and figures in this way: images include any products of clinical diagnostics, including photographs, micrographs, electrocardiogram rhythm images, and other products of diagnostic imaging, while figures are all other images, generally including data plots and illustrations.
- Provide evidence of relationships to other CCRs. This field may include identifiers (e.g., PMIDs) of other reports in the data set cited by or referencing this report.
- Provide evidence of relationships to clinical trials. This field may include identifiers of clinical trials citing this CCR. Identify trials by their ClinicalTrials.gov identifiers, preceded by NCT, or other stable identifier.
- Include database crosslinks corresponding to this document, including identifiers, preferably as database names and stable URLs.
4. Acknowledgements
Notes: Values in this category identify document-level features yet have little consistent structure across publications. They provide details regarding the organizations providing support for a CCR and related work. This category also includes a field for the total count of references cited by an article: this is intended to provide a rough metric of the degree to which a document has conceptual relationships with other biomedical documents of any type. Within the four data types in this section, provide the following.
- Specify all funding sources supporting the work and corresponding PI as well as relevant award numbers. The first value, Funding Source, should include the names of all organizations providing financial support for the work.
- Separate organizations with semicolons and spaces, e.g., National Institutes of Health/National Cancer Institute; DOE; Smith-Park Foundation.
- For the following value, Award Number, specify any award numbers or specific designations provided along with the recipients of the awards, where appropriate, as initials of the recipients in parentheses, e.g., R01HL123123 (to JP), NS12312 (to JP, JS), research training fellowship (to JS). Authors may explicitly state that no corresponding information is available (e.g., “no funding was received”); in these cases, use the text provided by the authors as the Funding Source value. Otherwise, the value should be NA.
- Specify disclosures/conflicts of interest as specified by the authors, e.g., JP is a consultant for DrugCo. Authors may explicitly state that no corresponding information is available (e.g., “no conflict of interest is declared”); in these cases, use the text provided by the authors as the Disclosures/Conflict of Interest value. Otherwise, as above, the value should be NA.
- Specify a numerical count of all references cited by the document, not including those provided in any supplementary material. No reference text should be included in this field.
Results
An example of the annotation process is shown in Figure 2. This case22 describes a presentation of infection by the bacterial pathogen Burkholderia thailandensis. For reference, the relevant portion of this CCR is provided in plain text format in Supplementary File 1; some research findings are also presented in this report and are included for comparison. In practice, converting reports provided in HTML or PDF format to plain text may improve the efficiency and ease of metadata extraction.
Examples of two sets of completed CCR metadata annotations are provided in Table 2. The first of these examples is mock data to illustrate the ideal format of each value, while the second example contains values extracted from a published CCR on a rare condition, acrodermatitis enteropathica23.
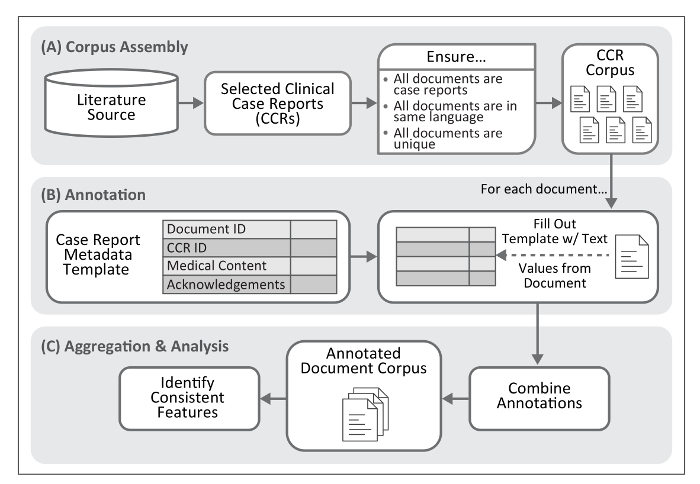
Figure 1. Workflow for Case Report Annotation. The protocol described here provides a method for identification of textual features frequently present within clinical case reports. This process requires assembly of a document corpus. The product of the annotation process, once aggregated into a single file, permits identification of text features associated with medical concepts and their descriptions within case reports. Please click here to view a larger version of this figure.

Figure 2. Identification of Concept-Specific Text in a Clinical Case Report. Beginning with the text of a case report, a manual annotator may progress through the document, identifying segments of text corresponding to each component of the metadata template. Identification features are highlighted in blue. Text corresponding to medical concepts are in red and labeled with their type; all highlighted text in the third column refers to the Pathology type. Please click here to view a larger version of this figure.
| Category | Description | ICD-10 Chapter | ICD-10 Code Range |
| cancer | Any type of cancer or malignant neoplasm. | II | C00-D49 |
| nervous | Any disease of the brain, spine, or nerves. | VI | G00-G99 |
| cardiovascular | Any disease of the heart or vascular system. Does not include hematological diseases. | IX | I00-I99 |
| musculoskeletal and rheumatic | Any disease of the muscles, skeletal system, joints, and connective tissues. | XIII | M00-M99 |
| digestive | Any disease of the gastrointestinal tract and digestive organs, including the liver and pancreas. | XI | K00-K95 |
| obstetrical and gynecological | Any disease relating to pregnancy, childbirth, the female reproductive system, or the breasts. | XIV; XV | O00-O9A; N60-N98 |
| infectious | Any disease causes by infectious microorganisms. | I | A00-B99 |
| respiratory | Any disease of the lungs and respiratory tract. | X | J00-J99 |
| hematologic | Any disease of the blood, bone marrow, lymph nodes, or spleen. | III | D50-D89 |
| kidney and urologic | Any disease of the kidneys or bladder, including the ureters, as well as the male reproductive organs, including the prostate. | XIV | N00-N53; N99 |
| endocrine | Any disease of the endocrine glands, as well as metabolic disorders. | IV | E00-E89 |
| oral and maxillofacial | Any condition involving the mouth, jaws, head, face, or neck. | XI; XIII | K00-K14; M26-M27 |
| eye | Any condition involving the eyes, including blindness. | VII | H00-H59 |
| otorhinolaryngologic | Any condition of the ear, nose, and/or throat. | VIII | H60-H95; J30-J39 |
| skin | Any disease of the skin. | XII | L00-L99 |
| rare | A special category reserved for reports of rare diseases, defined as those impacting fewer than 200,000 individuals in the United States (see https://rarediseases.info.nih.gov/diseases) | NA | NA |
Table 1. Disease Categories for Document Annotation. The categories listed here are those to be used for the Disease System data type in the document metadata template. As each disease presentation may involve multiple organ systems or etiologies, a single clinical case report may correspond to multiple categories. These categories largely follow those used to differentiate sections of the International Statistical Classification of Diseases and Related Health Problems, revision 10 (ICD-10) code system: corresponding ICD-10 chapters and code ranges are provided. Some categories, such as that for oral and maxillofacial disease, correspond to multiple sections of the ICD-10 system.
| Data Type | Example #1 | Example #2 (Cameron and McClain 1986) |
| Document and Annotation Identification | ||
| Internal ID | CCR005 | CCR2000 |
| Annotation Date | Mar 2 2018 | Mar 1 2018 |
| Case Report Identification | ||
| Title | A case of endocarditis. | Ocular histopathology of acrodermatitis enteropathica. |
| Authors | Grant AB;Chang CD | Cameron JD;McClain CJ |
| Year | 2017 | 1986 |
| Journal | World Journal of Medicine and Case Reports | British Journal of Ophthalmology |
| Institution | Department of Medicine, Division of Cardiology, First General Hospital, Boston, Massachusetts, USA | Department of Ophthalmology, University of Minnesota Medical School, Minneapolis, Minnesota 55455 |
| Corresponding Author | Grant AB | Cameron JD |
| PMID | 25555555 | 3756122 |
| DOI | 10.1011/wjmcr.2017.11.001 | NA |
| Link | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9555555/ | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1040795/ |
| Language | English | English |
| Medical Content | ||
| Key Words | brucellosis; endocarditis; mitral valve | NA |
| Demography | 37-year-old male | male child |
| Geographic Locations | Florida; Rio de Janeiro, Brazil | NA |
| Life Style | smoker; drinks alcohol occasionally | NA |
| Family History | third of five children of consanguineous parents; younger brother has chronic eczema | NA |
| Social History | construction worker | NA |
| Medical/Surgical History | history of fatigue | 8 pound 9 ounce (3884 g) product of an uncomplicated, full term pregnancy; in good health until age 1 month when he developed a blistering skin rash on his cheeks; rash spread to involve the skin around the eyes, nose, and mouth; skin lesions were also noted on the abdomen and extremities; diarrhoea and failure to thrive; skin biopsy at that time showed parakeratosis typical of acrodermatitis enteropathica; treated over the next six years with intermittent courses of broad spectrum antibiotics, breast milk, and diodoquin; partially responded; developed total alopecia, intermittent acrodermatitis, and intermittent diarrhoea with suboptimal weight gain; spasticity attributed to central nervous system involvement by the ae had developed by 8 months of age; several episodes of cardiopulmonary arrest at 11 months; lack of co-ordination of his vocal cords; tracheostomy; by age 18 months the child developed searching nystagmus associated with bilateral optic atrophy and slight attenuation of retinal vessels as well as signs of psychomotor retardation; bilateral keratoconjunctivitis; skin rash; second skin biopsy performed at age 3 again showed parakeratosis typical for ae; severe skin rash and diarrhoea; bilateral gross anterior corneal opacities were seen which had completely resolved by the time he was reexamined at age five; frequent infections including otitis media, urinary tract infections, and skin infections |
| Disease System | cardiovascular; infectious | digestive; skin; eye; rare |
| Signs and Symptoms | palpitations and dyspnea in the previous week; presented with lethargy, headache, and chills | severe blepharoconjunctivitis and bilateral anterior corneal vascularisation; severe skin rash and diarrhoea; gram-negative bacterial sepsis; skin lesions typical of acrodermatitis enteropathica, absence of thymic tissue, marked degeneration of the optic nerves, chiasm, and optic tracts and extensive cerebellar degeneration |
| Comorbidity | hypertension; hyperlipidemia | NA |
| Diagnostic Techniques and Procedures | Physical examination; electrocardiography; blood cultures | ocular examination; necropsy |
| Diagnosis | Brucella endocarditis | acrodermatitis enteropathica |
| Laboratory Values | increase in c-reactive protein (9 mg/dl); alkaline phosphatase (250 u/l) | NA |
| Pathology | Brucella melitensis was cultured from blood samples | right and left eyes were similar in appearance; corneal epithelium was reduced in thickness to one to three cell layers of flattened squamous epithelial cells over the entire surface of the cornea; all polarity of the epithelium was lost. bowman's membrane could be identified only in the periphery of the right cornea. no bowman's membrane could be identified in the left cornea. neither degenerative nor inflammatory pannus could be identified in either eye; extensive atrophy of the circular and oblique muscles of the ciliary body; some posterior migration of lens capsular epithelium and early cortical degenerative changes; extensive degeneration of the retinal pigment epithelium throughout the posterior pole; retina was attached and showed mild autolytic changes throughout; some preservation of rod and cone outer segments in the posterior pole, however, these structures were completely lost anterior to the equator; extensive loss of the ganglion cell and nerve fibre layers of both eyes; nearly complete atrophy of the disc and adjacent optic nerve |
| Pharmacological Therapy | gentamycin 240 mg/iv/daily | NA |
| Inverventional Therapy | prosthetic valve replacement | NA |
| Patient Outcome Assessment | recovery was uneventful; discharged home | died in 1971 (age 7) |
| Diagnostic Imaging/Videotape Recording | 2;1;0;1 | 7;0;0;0 |
| Relationship to Other Case Reports | 5555555 | 23430849 |
| Relationship with Clinial Trial | NCT05555123 | NA |
| Crosslink with Database | MedlinePlus Health Information: https://medlineplus.gov/ency/article/000597.htm | HighWire - PDF: http://bjo.bmj.com/cgi/pmidlookup?view=long&pmid=3756122; Europe PubMed Central: http://europepmc.org/abstract/MED/3756122; Genetic Alliance: http://www.diseaseinfosearch.org/result/143 |
| Acknowledgements | ||
| Funding Source | National Institutes of Health/National Heart, Lung, and Blood Institute | The Minnesota Lions Club; Research to Prevent Blindness; Veterans Administration; Office of Alcohol and Other Drug Abuse Programming of the State of Minnesota |
| Award Number | R01HL123123 (to AG) | NA |
| Disclosures/Conflict of Interest | Dr. Grant is a paid spokesperson for DrugCo. | NA |
| References | 4 | 27 |
Table 2. Standardized Metadata Template for Clinical Case Reports, with Example Annotations. A set of features common to clinical case reports and facilitating their concept-level annotations is shown here. This template is arranged into three primary sections: Identification, Medical Content, and Acknowledgments, denoting the purpose and additional value afforded by each type of case report feature. This table contains two sets of example annotations, one of a fictionalized case report, and another set derived from a report on the condition acrodermatitis enteropathica23.
Supplementary File 1. Text of a clinical case report (Chang et al. 2017). Please click here to download this file.
Discussion
Implementation of a standardized metadata template for CCRs can make their content more FAIR, expand their audience, and extend their applications. Following the traditional use of CCRs as educational tools in medical communications, healthcare trainees (e.g., medical students, interns, and fellows), and biomedical researchers may find that summarized case report contents enable more rapid comprehension. The greatest strength of metadata standardization with CCRs, however, is that indexing these data transforms otherwise isolated observations into interpretable patterns. The protocol provided here can serve as the first step in a workflow for working with CCRs, whether this workflow consists of epidemiological analysis, post-marketing drug or treatment surveillance, or broader surveys of pathogenesis or therapeutic efficacy. Structured features identified within CCRs can provide a useful resource for researchers focusing on disease presentations and treatments, particularly for rare conditions. Clinical researchers may find data on past treatment regimens to analyze recorded symptoms or side effects and degree of improvement under previous standards of care. The data may also drive broader analyses of a new treatments based on efficacy, lack of adverse effects or toxicity, or on drug targeting differences in gender, age group, or genetic background.
The benefits provided by structured metadata are similarly applicable to computational workflows designed to parse or model medical language. Structured CCR features may also provide evidence of areas where report authors may provide more easily machine-readable (and in some cases, human-readable) content. Variance among CCRs can result from a lack of explicitly provided observations: e.g., a patient's exact age may not be specified. Similarly, clinicians may not mention tests if the diagnostics or their results were considered trivial. By providing examples of gaps necessary for in-depth analysis, enforcing structure on CCRs highlights potential improvements. In a broader perspective, a greater availability of structured text data from medical documents supports natural language processing (NLP) efforts to learn from big data in healthcare24,25.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported in part by National Heart, Lung, and Blood Institute: R35 HL135772 (to P. Ping); National Institute of General Medical Sciences: U54 GM114833 (to P. Ping, K. Watson, and W. Wang); National Institute of Biomedical Imaging and Bioengineering: T32 EB016640 (to A. Bui); a gift from the Hoag Foundation and Dr. S. Setty; and the T.C. Laubisch endowment at UCLA (to P. Ping).
Materials
| Name | Company | Catalog Number | Comments |
| A corpus of clinical case reports | n/a | n/a | Full texts of case reports may be accessed through PubMed (e.g., using the search query "Case Reports"[Filter]), other citation databases such as Europe PMC (https://europepmc.org/) or directly through publishers. |
References
- Ban, T. A. The role of serendipity in drug discovery. Dialogues in Clinical Neuroscience. 8 (3), 335-344 (2006).
- Cabán-Martinez, A. J., García-Beltrán, W. F. Advancing medicine one research note at a time: the educational value in clinical case reports. BMC Research Notes. 5 (1), 293(2012).
- Vandenbroucke, J. P. In Defense of Case Reports and Case Series. Annals of Internal Medicine. 134 (4), 330(2001).
- Bayoumi, A. M. The storied case report. Canadian Medical Association Journal. 171 (6), 569-570 (2004).
- Pasteur, L. Méthode pour prévenir la rage après morsure. Comptes rendus de l'Académie des Sciences. 101, 765-774 (1885).
- Pearce, J. Louis Pasteur and Rabies: a brief note. Journal of Neurology, Neurosurgery & Psychiatry. 73 (1), 82-82 (2002).
- Keefer, C. S., Blake, F. G., Marshall, E. K. J., Lockwood, J. S., Wood, W. B. J. PENICILLIN IN THE TREATMENT OF INFECTIONS. Journal of the American Medical Association. 122 (18), 1217(1943).
- Akers, K. G. New journals for publishing medical case reports. Journal of the Medical Library Association JMLA. 104 (2), 146-149 (2016).
- Wilkinson, M. D., et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data. 3, 160018(2016).
- Beeler, G. W. HL7 Version 3-An object-oriented methodology for collaborative standards development. International Journal of Medical Informatics. 48 (1-3), 151-161 (1998).
- HL7 FHIR Release 3 (STU; v3.0.1-11917). , Available from: http://hl7.org/implement/standards/fhir/ (2018).
- McDonald, C. J. LOINC, a Universal Standard for Identifying Laboratory Observations: A 5-Year Update. Clinical Chemistry. 49 (4), 624-633 (2003).
- CDC/National Center for Health Statistics ICD-10-CM Official Guidelines for Coding and Reporting. , Available from: https://www.cdc.gov/nchs/data/icd/10cmguidelines_fy2018_final.pdf (2017).
- Rajeev, D., et al. Development of an electronic public health case report using HL7 v2.5 to meet public health needs. Journal of the American Medical Informatics Association. 17 (1), 34-41 (2010).
- Biesecker, L. Mapping phenotypes to language: a proposal to organize and standardize the clinical descriptions of malformations. Clinical Genetics. 68 (4), 320-326 (2005).
- Riley, D. S., et al. CARE guidelines for case reports: explanation and elaboration document. Journal of Clinical Epidemiology. 89, 218-235 (2017).
- Cohen, K. B., et al. Coreference annotation and resolution in the Colorado Richly Annotated Full Text (CRAFT) corpus of biomedical journal articles. BMC Bioinformatics. 18 (1), 372(2017).
- Sun, W., Rumshisky, A., Uzuner, O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. Journal of the American Medical Informatics Association. 20 (5), 806-813 (2013).
- Savova, G. K., et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 17 (5), 507-513 (2010).
- Soysal, E., et al. CLAMP - a toolkit for efficiently building customized clinical natural language processing pipelines. Journal of the American Medical Informatics Association. 25 (3), 331-336 (2018).
- Bender, D., Sartipi, K. HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems. , 326-331 (2013).
- Chang, K., et al. Human Infection with Burkholderia thailandensis, China, 2013. Emerging Infectious Diseases. 23 (8), 1416-1418 (2013).
- Cameron, J. D., McClain, C. J. Ocular histopathology of acrodermatitis enteropathica. British Journal of Ophthalmology. 70 (9), 662-667 (1986).
- Maddox, T. M., Matheny, M. A. Natural Language Processing and the Promise of Big Data. Circulation: Cardiovascular Quality and Outcomes. 8 (5), 463-465 (2015).
- Kreimeyer, K., et al. Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. Journal of Biomedical Informatics. 73, 14-29 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved