Method Article
High-resolution Patterned Biofilm Deposition Using pDawn-Ag43
In This Article
Summary
We demonstrate a method for depositing Escherichia coli bacterial biofilms in arbitrary spatial patterns with a high resolution using optical stimulation of a genetically encoded surface-adhesion construct.
Abstract
Spatial structure and patterning play an important role in bacterial biofilms. Here we demonstrate an accessible method for culturing E. coli biofilms into arbitrary spatial patterns at high spatial resolution. The technique uses a genetically encoded optogenetic construct—pDawn-Ag43—that couples biofilm formation in E. coli to optical stimulation by blue light. We detail the process for transforming E. coli with pDawn-Ag43, preparing the required optical set-up, and the protocol for culturing patterned biofilms using pDawn-Ag43 bacteria. Using this protocol, biofilms with a spatial resolution below 25 μm can be patterned on various surfaces and environments, including enclosed chambers, without requiring microfabrication, clean-room facilities, or surface pretreatment. The technique is convenient and appropriate for use in applications that investigate the effect of biofilm structure, providing tunable control over biofilm patterning. More broadly, it also has potential applications in biomaterials, education, and bio-art.
Introduction
Biofilms are surface-attached communities of microbes, and are well-known for their strong structure-function coupling. Spatial geometry and patterning of biofilms play an important role in overall community function (and vice versa)1. The small length scales involved in biofilm structure—on the order of tens of microns2—make tunable and convenient control of biofilm patterning a challenging problem. Here we demonstrate a protocol that allows for biofilms to be precisely patterned in arbitrary geometries, based on optical illumination.
The protocol presented here uses pDawn-Ag433, an optogenetic construct that couples biofilm formation in E. coli bacteria to optical illumination by driving the expression of Ag43 (an adhesin gene responsible for surface adhesion and biofilm formation) under the control of pDawn4 (a transcriptional regulator controlled by optical illumination). The method is convenient to use and can pattern biofilms on various surface environments, including enclosed (transparent) culture chambers. Compared to existing cell deposition methods, such as droplet-based deposition5 or surface prepatterning/treatment6, pDawn-Ag43 does not require microfabrication or clean-room facilities and does not require materials beyond those available to a typical microbiology laboratory. It is able to pattern with a spatial resolution below 25 μm, approaching the spatial dimensions of microcolonies in naturally existing biofilms2. Overall, this technique provides the ability to manipulate biofilm structure, which then opens many avenues to study microecology in bacterial communities7. Additionally, patterned biofilms may provide a convenient platform upon which to engineer useful biomaterials8,9. In this paper, we discuss the basic protocol required for patterning biofilms using pDawn-Ag43 and address potential modifications and troubleshooting related to the method.
Protocol
1. Preparation of pDawn-Ag43 Bacterial Strains
- Transform pDawn-Ag43 into an E. coli strain of interest (Figure 1).
- Grow a cloning strain hosting pDawn-Ag43 plasmid (obtainable from a plasmid repository) by inoculating the strain in LB broth supplemented with 50 μg/mL spectinomycin (LB+spec) in a culture tube (overnight in a shaking incubator at ~250 rpm, 37 °C). Then, use a miniprep kit to harvest the purified pDawn-Ag43 plasmid10.
- Choose an E. coli strain of interest to be patterned. Thus far, pDawn-Ag43 has been verified to work in MG16553 and BW25113.
- Use an established protocol to generate competent (e.g., chemically competent11 or electrocompetent12) stocks of the chosen E. coli strain (e.g., MG1655).
- Transform pDawn-Ag43 plasmid into the competent bacteria11,12, followed by 1 h recovery, and plate on LB+spec agar plates. Allow colonies to grow overnight at 37 °C.
- Store the pDawn-Ag43 transformed strains.
- Inoculate a single colony of pDawn-Ag43 from an LB+spec agar plate into LB+spec broth in a culture tube and grow it in a shaking incubator (at ~250 rpm, 37 °C) to exponential phase (OD600 ~0.4 - 0.8).
- Prepare stock to store the pDawn-Ag43 transformed strain at -80 °C long-term by mixing 1 mL of culture with 1 mL of 50% sterile glycerol in a cryo-tube to obtain a 25% glycerol freezer stock. Store this in a -80 °C freezer.
- If additional plasmids (e.g., fluorescent reporter plasmids) need to be transformed, create competent cells11,12 from the pDawn-Ag43 transformed strain and repeat the transformation process11,12 for additional plasmids as needed.
NOTE: If using electroporation8, it is possible to cotransform multiple plasmids (including pDawn-Ag43) simultaneously; however, a simultaneous transformation is not recommended using chemical transformation methods, as the large size (> 10 kB) of pDawn-Ag43 means transformation efficiencies are reduced.
2. Preparation of the Projector Optical Set-up for Illuminating Bacteria
- Obtain a bacterial incubator with non-transparent walls and a hole for passing cables, a portable projector capable of fitting and functioning inside the bacterial incubator, and a laptop equipped with software for presentation-projection (see Table of Materials). When choosing the projector and incubator, ensure that the minimal focus distance of the projector is less than the interior height of the incubator.
- Place the projector inside the incubator at the bottom, with the aperture pointing directly upward, at the ceiling, where the biofilm culture chamber is attached (Figure 2).
- Fix the projector in place by constructing a set-up with an optical breadboard base connected to a vertical post, in turn connected to a horizontal post which screws into the projector (Figure 2). Note that this set-up can be modified depending on the projector/available parts. Ultimately, the key requirement is that the projector is solidly fixed near the bottom of the incubator, with the aperture pointing upward.
- Connect the laptop via the display cable (e.g., HDMI) to the projector inside the incubator.
- If the surfaces—especially the ceiling—of the interior of the incubator are reflective (e.g., metal polished surface), cover them with dark matte surfaces to minimize reflections.
- Use tape to attach an empty 'dummy' culture dish to the ceiling of the incubator. Note that, as with the projector, there are multiple acceptable ways to attach a culture dish; ensure that the transparent bottom surface of the culture chamber, where illumination occurs, is not covered.
- Adjust the focus on the projector by turning the focusing knob so that the focusing plane coincides with the bottom surface of the biofilm culture dish (e.g., a well plate) attached to the ceiling of the incubator (Figure 2). The projector should illuminate a sharp, non-blurry image onto the bottom of the culture dish. Remove the 'dummy' culture dish once the projection has been optimized.
- Using laptop software, direct the projector to illuminate the full field of view with maximum blue illumination (e.g., RGB = [0, 0, 255]) by presenting a full blue slide.
- Measure the illumination intensity of the projector using an optical power meter by placing the photodetector head at the incubator ceiling and reading the intensity on the corresponding power meter calibrated to light wavelength 460 nm. Follow the instructions on connecting and calibrating the photodetector for the specific power meter used.
- Reduce ambient light (e.g., turn off room lights, or place the incubator away from light sources) as much as possible prior to making illumination intensity measurements.
- Adjust the illumination intensity using an adjustable neutral density filter placed at the projector aperture. Rotate the filter to adjust the illumination intensity measured by the power meter, until the illumination intensity in the center of the blue-light projected region reads 50 μW/cm2.
NOTE: While it is possible to adjust the illumination intensity by using lower blue RGB values on the software end, using a filter while maximizing the blue RGB value has the advantage of maximizing the optical contrast of the system between illuminated vs. dark regions. - Draw arbitrary patterns using the presentation/projector software on the laptop and display these patterns on the ceiling of the incubator, using the projector.
NOTE: Biofilm-forming regions should be drawn with maximum blue illumination (e.g., RGB = [0, 0, 255]), non-biofilm-forming regions with no illumination (e.g., RGB = [0, 0, 0]).
3. Culturing Patterned Biofilms
- Prior to illumination, prepare the pDawn-Ag43 bacteria, starting from the glycerol freezer stock, so that they are reliably induced at the late exponential growth phase for illumination (Figure 3A).
- Streak a pDawn-Ag43 strain from glycerol stock onto LB+spec agar plates. Allow colonies to grow overnight (37 °C).
NOTE: From here until the illumination culture step, ensure the cells stay in the dark as much as possible—brief periods at ambient illumination (e.g., for subdilution) are acceptable. - Inoculate a colony of pDawn-Ag43 bacteria from an agar plate in LB+spec broth and grow the culture overnight to stationary phase (in a shaking incubator at ~250 rpm, 37 °C, for ~16 h).
- Subdilute the culture at a ratio of 1:1,000 with LB+spec broth (e.g., add 1 μL of overnight culture to 1 mL of fresh LB+spec).
- Allow the subdiluted culture to grow until late exponential/early stationary phase (OD ~1.0, ~6 h in a shaking incubator at ~250 rpm, 37 °C).
- While waiting for the culture to grow, prepare M63 media with 1x M63 salts, 1 mM MgSO4, 0.2% glucose, 0.1% casamino acids, and 50 μg/mL spectinomycin in water (ensure the constituent parts are sterile).
- Subdilute the late-exponential-phase culture at a ratio of 1:100 with M63 media supplemented with 50 μg/mL spectinomycin. Then, introduce the dilution into a biofilm culture dish (e.g., pipette the diluted sample into a well plate).
NOTE: The volumes required for this 1:100 subdilution depend on the culture dish being used (e.g., if using a 6-well polystyrene well plate [non-tissue-culture treated], a single sample would require adding 20 μL of culture to 2 mL of M63+spec, as the standard well volume of a 6-well plate is ~2 mL).
- Streak a pDawn-Ag43 strain from glycerol stock onto LB+spec agar plates. Allow colonies to grow overnight (37 °C).
- As the samples are now ready for illumination, tape the culture dish to the ceiling of the incubator, ensuring that the surface at the bottom of the dish is transparent for illumination from below by the projector.
NOTE: It is important to ensure that the ceiling of the incubator is not reflective, to minimize stray illumination. Stray illumination can also be reduced by using black-walled plates as culture dishes, although this is not strictly necessary—if using such plates, ensure the bottom surface is transparent. - Allow the biofilms to culture in the incubator overnight (16 h with no shaking, at 37 °C). Note that some projectors become less reliable at higher temperatures. If that is the case, culture at lower temperatures (e.g., 30 °C), keeping in mind that the incubation time may need to be increased, depending on the E. coli strain.
4. Imaging Patterned Biofilms
- After the overnight growth of the biofilm samples, remove the culture dish from the incubator. The dish will have biofilm bacteria attached to its bottom surface where it has been illuminated, as well as planktonic bacteria dispersed in the liquid media above.
- Discard the planktonic cells by removing the liquid media from the culture dish (e.g., by gently aspirating with a pipette).
- Rinse the sample 2x with a phosphate-buffered saline (PBS) solution to remove the remaining planktonic cells (Figure 3B) by gently pipetting in PBS, followed by aspiration.
- If the cells are fluorescently tagged, directly image the samples using fluorescence microscopy13 (e.g., wide-field13, 3-D confocal14, etc.).
NOTE: Fluorescent biofilms can also be preserved using a self-hardening mounting medium. Apply one drop of mounting media to a biofilm sample, cover it with a glass coverslip, taking care not to capture any bubbles underneath, and allow it to harden overnight before imaging. - If the bacterial cells used are not fluorescently tagged, apply the crystal violet stain technique15 (Figure 3B) to enhance biofilm contrast prior to imaging.
5. Protocol Modifications/Alternatives
- Grow pDawn-Ag43 bacteria on different surfaces.
- Put glass or poly-dimethyl-siloxane (PDMS) coupons (e.g., coverslips or thin strips of PDMS) into well plates prior to the addition of a bacterial sample/illumination, and follow the same protocol as before to pattern pDawn-ag43 biofilms on glass and PDMS.
- Grow pDawn-Ag43 bacteria inside a transparent, enclosed culture chamber.
- Generate a culture chamber mold for PDMS.
- For a basic culture chamber, generate mold by attaching a hard, rectangular prism to a flat surface (the prism will become a cavity in the PDMS that serves as the culture chamber once the mold is cast). Note that more intricate culture chamber molds can be fabricated using soft lithography16.
- Cast a PDMS cavity by pouring PDMS into the mold and allowing it to cure (for a detailed soft lithography protocol, see JoVE's Science Education Database16).
- After curing, trim any excess PDMS, punch inlet/outlet channels into the cavity/culture chamber and bond the cavity to a flat surface (e.g., glass/polystyrene) by firmly pressing the PDMS onto the flat surface, leaving the cavity between the surface and the PDMS ceiling as the biofilm culture chamber.
NOTE: More permanent bonding based on plasma treatment16 can also be used, but the chips will then not be reusable. - Follow the culture protocol as before, using a syringe with blunt tip needles (instead of a pipette) to introduce the bacterial sample into the culture chamber/rinse with PBS buffer (Figure 3C).
- If using a temporarily bonded cavity, use only negative pressure to pull liquid into/out of the chamber, to ensure that the cavity does not become unbonded from the underlying glass/polystyrene surface.
- Generate a culture chamber mold for PDMS.
- Grow pDawn-Ag43 bacteria using a film photomask for structured illumination.
- Design a biofilm pattern using CAD software compatible with a film photomask printer/print service. The photomask film design should be clear in regions where the biofilm is meant to be printed, and black/opaque elsewhere. When complete, send the photomask file to the printer/printing service and await the return of the physical photomask.
- Instruct the projector to illuminate a full field of view with maximum blue illumination (e.g., RGB = [0, 0, 255]) using laptop software.
- Cut out a region of interest from the larger film photomask, and tape it directly to the bottom of the biofilm culture dish prior to introducing the bacterial sample for overnight illumination (Figure 3D). Culture the biofilms as before and remove the photomask after culturing, prior to imaging.
Results
As seen in Figure 4A, pDawn-Ag43 bacteria were used to generate biofilms patterned in polystyrene well plates with projector illumination (the projector was set to illuminate a polka-dot pattern), imaged through brightfield microscopy with crystal violet stain as a contrast agent, and fluorescence microscopy using red-fluorescent-protein-expressing bacteria. Fluorescent biofilm samples can also be imaged using confocal microscopy14 to obtain images of the biofilm in 3-D (Figure 4B). In Figure 4C, we illustrate the high-resolution patterning possible by using a film photomask to provide patterned illumination to the biofilm sample. Finally, in Figure 4D and 4E, we demonstrate examples of patterning on glass and PDMS surfaces, as well as enclosed PDMS culture chambers—these illustrate the different types of environments where pDawn-Ag43 patterning can be applied.
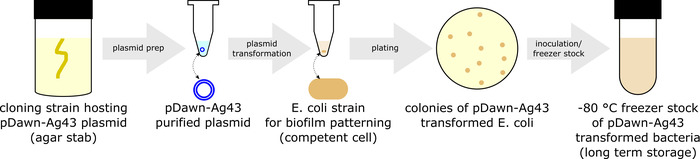
Figure 1: Preparation of pDawn-Ag43 bacteria (protocol section 1). Preparing pDawn-Ag43 bacteria capable of light-regulated biofilm formation involves purifying pDawn-Ag43 plasmid from a host cloning strain, transforming it into an E. coli strain of interest, and creating bacterial freezer stock for long-term storage. Please click here to view a larger version of this figure.
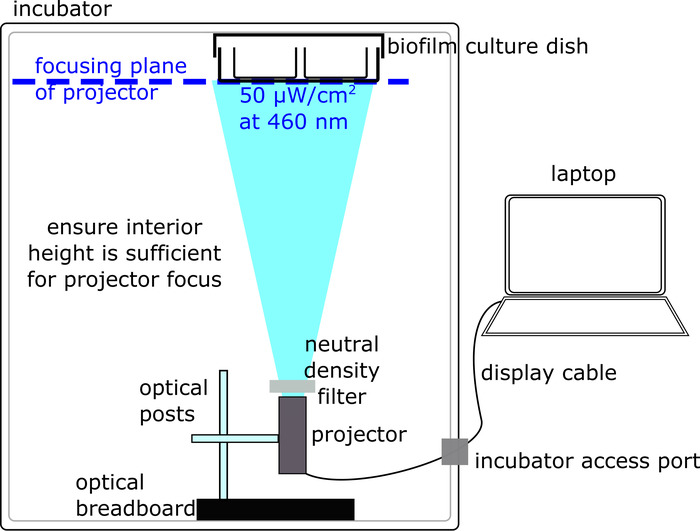
Figure 2: Preparation of an optical set-up for biofilm sample illumination (protocol section 2). The optical set-up is housed inside a bacterial incubator and consists of a computer-connected projector illuminating a biofilm sample. Please click here to view a larger version of this figure.
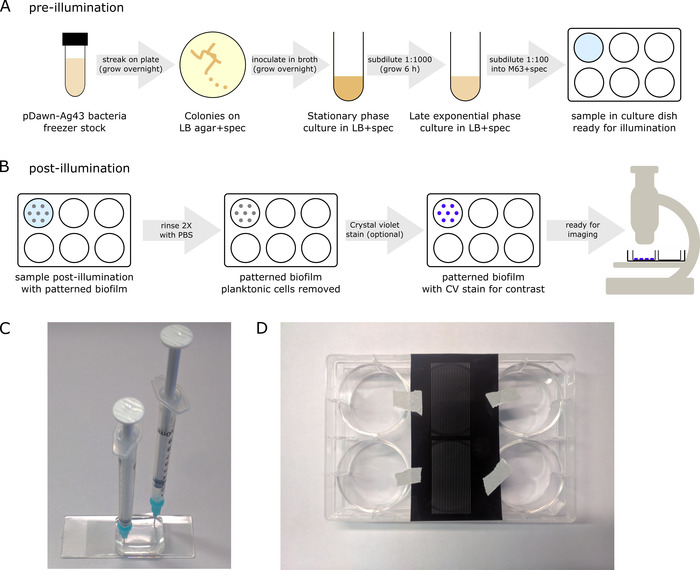
Figure 3: Culture protocol for patterning biofilms (protocol section 3). (A) Prior to illumination, pDawn-Ag43 bacteria are prepared prior to patterning such that they are reliably induced at the proper growth phase. (B) After overnight illuminated growth, a patterned biofilm will be present at the bottom of the culture dish, along with planktonic cells in the liquid media, and after some further processing, the biofilm is ready for imaging. (C) As an alternative to well plates, biofilms can be cultured in enclosed culture chambers such as a molded PDMS cavity. In this case, syringes attached to blunt tip needles can be used to introduce the sample and flush liquids out of the chamber. (D) As an alternative to projector-based illumination patterns, patterns can also be generated by taping film photomasks directly to the bottom of biofilm culture chambers. In this case, the projector should be set up to illuminate blue light across the full field. Please click here to view a larger version of this figure.
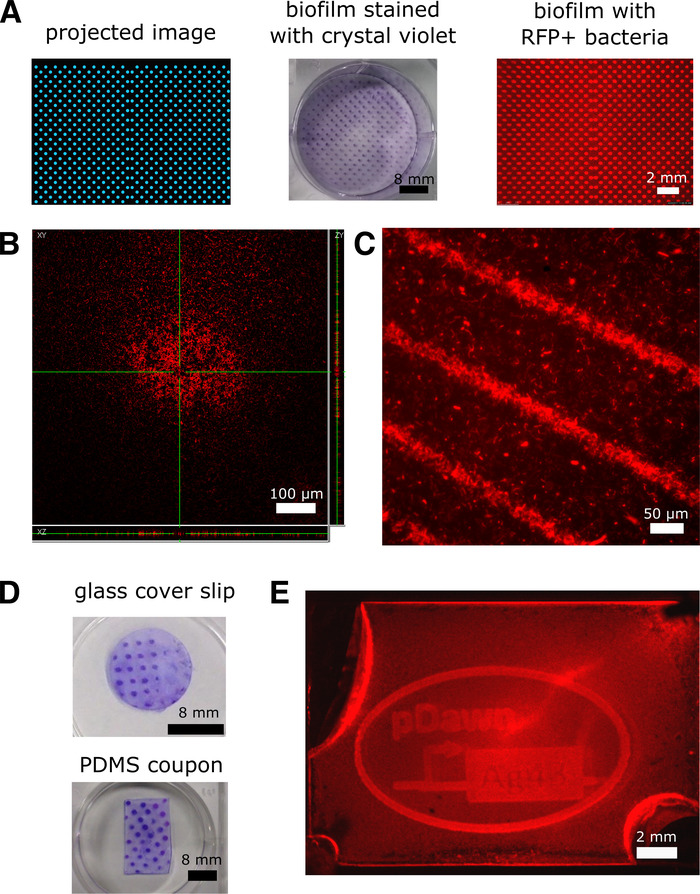
Figure 4: Representative results of biofilms patterned using pDawn-Ag43. All results were obtained using an MG1655 E. coli host strain. (A) pDawn-Ag43 bacteria were used to generate biofilms patterned in polystyrene well plates with projector illumination (the projector was set to illuminate a polka-dot pattern), imaged through brightfield microscopy with crystal violet stain as a contrast agent, and fluorescence microscopy using red-fluorescent-protein-expressing bacteria. (B) Fluorescent biofilm samples are imaged with confocal microscopy to obtain 3-D images of the biofilm. (C) High-resolution biofilms can be patterned with a film photomask to provide patterned illumination to the biofilm sample. (D) Biofilms can be patterned on glass and PDMS surfaces. (E) Biofilms can be patterned in enclosed culture chambers. This figure has been adapted from previous work3. Please click here to view a larger version of this figure.
| Problem | Potential causes/solutions |
| Tranforming pDawn-Ag43 into host strain - no colonies | Low plasmid concentration - check plasmid concentration on spectrometer. A typical miniprep of pDawn-Ag43 should yield at least 100 ng/μL; use up to 10-100 ng for transformation |
| Check/remake competent cells: competent cells should have transformation efficiency at least 10^6 cfu/µg verified using a standard plasmid such as pUC19 - if not, remake competent cells | |
| Wrong (level of) antibiotic on LB agar plate - make sure to use 50 μg/mL spectinomycin for selection | |
| Projector illumination turns off / inconsistent overnight | Disable problematic software such as: automatic overnight software/OS updating, night-time blue-light filter |
| Projector may be overheating - set incubator to lower temperature while projector is turned on (e.g. 30 °C instead of 37 °C) - note projector as heat source can overheat incubator beyond set point | |
| Remove unnecessary sources of humidity from incubator, as these may affect projector electronics | |
| No/low levels of biofilm formed after overnight illumination, no planktonic cells growth either (i.e. liquid is clear) | Wrong (level of) antibiotic - make sure to use 50 μg/mL spectinomycin |
| Check everything is added to M63 recipe properly | |
| No/low levels of biofilm formed after overnight illumination, only planktonic cells (i.e. liquid is cloudy) | Check light level, projector should be illuminating blue light at 50 μW/cm^2 at 460 nm wavelength |
| Try letting bacteria grow for shorter/longer time after 1:1000 LB subdilution step prior to adding to M63 | |
| Restreak bacteria on LB plate, start from fresh colony to generate overnight stationary phase culture | |
| Ensure projector is working consistently overnight - see point above | |
| Fuzzy biofilm patterns, high levels of background noise | Reduce stray light from optical illumination system, cover reflective surfaces on interior of incubator |
| Consider using photomask-based (as opposed to projector-based) structured illumination | |
| Check projector is properly focused at the bottom surface of the biofilm culture chamber |
Table 1: Common troubleshooting issues.
Discussion
In light of the need for research tools that allow for biofilm structure control, we have presented an easy-to-use protocol for patterning bacterial biofilms using the pDawn-Ag43 optogenetic construct. With this technique, E. coli biofilms can be optically patterned on various surface environments, including enclosed chambers, with a spatial resolution below 25 μm.
Overall, this protocol can be broken down into four main sections: (1) the preparation of the pDawn-Ag43 bacteria, (2) the preparation of the optical and culture set-up hardware, (3) the pre-illumination bacterial growth steps, and (4) the post-illumination rinses and imaging.
The critical part of section 1 is the successful transformation of pDawn-Ag43 plasmid into the E. coli strain of interest. This is facilitated by isolating high-quality purified plasmid and generating high-quality competent cells for transformation (Table 1, troubleshooting).
The critical part of section 2 is the optimization of the projector set-up so that the illumination intensity is adjusted to 50 μW/cm2 at the 460-nm wavelength, and the projector is properly focused at the biofilm sample height. Note that in this protocol, we describe an inverted illumination set-up where the projector shines light from below, upward toward the biofilm sample. The advantage of this set-up is that the light only needs to travel through the bottom of the culture dish before reaching the biofilm formation surface. Illumination from above means that the light would have to travel through the liquid media above the biofilm surface, which, during the course of the growth, gets cloudy with planktonic cells. In addition to these concerns, it is also important to minimize stray light in the optical set-up as much as possible, for example, by covering up reflective surfaces on the interior of the incubator—this helps to obtain sharper patterned biofilms. On a related note, sharper biofilm patterns can also be obtained by using a photomask to control illumination patterning (Figure 3D, Figure 4C). Common issues requiring troubleshooting include projector reliability issues at higher temperatures (e.g., 37 °C), which can be minimized by incubating the biofilm growth at lower temperatures (e.g., 30 °C), as well as computer software that causes operating system updates or blue light filtering during overnight growth (Table 1). It is also important to note that, depending on the projector and incubator model used, it is also possible that heat generated from the projector will result in a higher interior temperature than the incubator set temperature, which may need to be corrected.
The critical part of section 3 is obtaining reliable and repeatable bacterial samples before they are induced by illumination. For this reason, it is recommended to obtain clonal colonies of pDawn-Ag43 bacteria by streaking them out on an agar plate and then using the liquid culture steps to ensure that the bacteria are illuminated/induced at the late exponential growth phase in a repeatable manner.
Finally, the critical part of section 4 is to thoroughly, but also gently, wash away the planktonic cells remaining after the biofilm patterning protocol; thus, it is recommended to perform multiple gentle rinse steps with PBS.
Compared to existing techniques for cell patterning5,6, optical biofilm patterning based on pDawn-Ag43 has a reasonably low barrier of entry to use, in that it does not require microfabrication, clean-room facilities, complex chemistry, or surface pretreatment, yet is still able to pattern with the high resolution (25 μm) typically associated with microfabrication techniques. The method extends previous work on bacterial photolithography for controlling gene expression17. Currently, pDawn-Ag43 plasmid is limited to E. coli, as it uses a pUC-based origin of replication, but pDawn and Ag43 are both compatible in other (Gram-negative) bacterial species. Genetic techniques are available for potentially introducing light-regulated biofilm formation to different bacterial species and represents a possible direction for future research. Another potential limitation of the technique is that it works by increasing biofilm formation in strains with weak native biofilm formation (e.g., MG1655 E. coli). However, strains with strong native biofilm formation have biofilms form regardless of illumination conditions, precluding patterned biofilm formation using pDawn-Ag43 as described here; yet optogenetic techniques may still prove applicable in regulating biofilm formation. We note that in other contexts, alternative methods of biofilm patterning may be available, such as via optical c-di-GMP modulation18.
Overall, pDawn-Ag43 based patterning will be appropriate for use in applications that investigate the effect of biofilm structure on function1 and, therefore, could benefit from tunable control over biofilm patterning—a particularly relevant example to highlight is the study of microbial ecology in biofilms2. Future directions include making patterned biomaterials8,9 and/or structured bacterial communities. Alternative applications of this accessible protocol also include bio-art19, given the clear aesthetic potential, as well as formal and informal life science education20,21,22. From an educational perspective, the protocol described here combines many relevant techniques (bacterial culture, transformation, optics/optogenetics) and is also modularly extendable (e.g., include microfluidics).
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank D. Glass, H. Kim, N. Cira, A. Choksi, S. Rajan, and B. Keys for their helpful suggestions and the Spormann lab for access to their confocal microscope. Furthermore, the authors acknowledge the support from Stanford Bio-X Bowes and NSERC PGS fellowships, the National Institute of Health (R21-AI-139941), and the American Cancer Society (RSG-14-177-01).
Materials
| Name | Company | Catalog Number | Comments |
| DH5alpha/pDawn-Ag43 | Addgene | 107742 | DH5alpha cloning strain hosting pDawn-Ag43 plasmid - plasmid needs to be moved to E. coli strain of interest prior to use |
| MG1655 E. coli | Coli Genetic Stock Center - Yale University | CGSC #6300 | MG1655 was used as E. coli strain of interest in this paper's representative results |
| RFP expression plasmid | iGEM biobricks | J04450-pSB4K5bb | Many options exist to obtain fluorescent bacteria - if using plasmid, ensure backbone does not conflict with colE1 ori of pDawn-Ag43 |
| Plasmid miniprep kit | Qiagen | 27104 | |
| LB broth powder | Affymetrix | 75852 | Add 20 g/L to water, autoclave, add 50 μg/mL spectinomycin to get sterile LB+spec |
| LB agar powder | Affymetrix | 75851 | Add 35 g/L to water, autoclave, add 50 μg/mL spectinomycin, pour into petri dishes to get sterile LB+spec plates |
| Petri dishes | Fisherbrand | 431760 | |
| Spectinomycin hydrochloride pentahydrate | abcam | ab141968 | Make 1000x stock 50mg/mL in water, filter sterilize and dilute into media as needed |
| Glycerol | Sigma-Aldrich | G5516 | Mix at 1:1 ratio with water, sterilize by autoclave or filter to obtain 50% glycerol |
| M63 media salts 5X solution | Bio-world | 705729 | Add cas-amino acids, glucose and MgSO4, bring to 1X salts concentration by adding sterile water |
| Casamino acids | Amresco | J851 | Make 20% stock in water, filter sterilize and add to M63 as supplement (final concentration 0.1%) |
| D-glucose | Sigma-Aldrich | G8270 | Make 20% stock in water, filter sterilize and add to M63 as supplement (final concentration 0.2%) |
| Magnesium sulfate | Sigma-Aldrich | M7506 | Make 1 M stock in water, autoclave and add to M63 as supplement (final concentration 1 mM) |
| Crystal violet | Acros organics | 212120250 | Dilute to 0.1% in water prior to use |
| Self-hardening mounting media (Shandon immumount) | Thermo Scientific | 9990402 | Use to preserve samples over long term for fluorescence imaging |
| Phosphate buffered saline (PBS) solution | Gibco | 10010023 | Can also use PBS prepared from powder / tablets |
| 6 well plate | Fisherbrand | 351146 | Used as biofilm culture dish for representative results |
| PDMS kit | Dow | SYLGARD 184 | Can be used to make enclosed microchamber cavities using soft lithography |
| 1 mL syringe | BD syringe | 309659 | For use with liquid handling with enclosed microchambers |
| Blunt tip needle | CML supply | 901-23-050 | Attaches to 1 mL syringe |
| Lab tape | Fisherbrand | 15-901 | Use to attach culture chamber to incubator ceiling |
| Bacterial incubator | Sheldon Manufacturing | SMI6 | Ensure interior height of incubator is tall enough to focus projector at the ceiling |
| Portable projector | Ivation | IV-PJ-PRO-4-1 | Many portable projector models exist, pDawn-Ag43 has been tested with multiple models including LED/laser based, with blue light channel ranging from 450-460nm central wavelength |
| Optical breadboard base | ThorLabs | MSB6 | Base for optical setup to hold projector - many other setups possible, just need to hold projector firmly at bottom of incubator, pointing upwards |
| Optical post | ThorLabs | TR8 | 2 posts needed - one to be set up vertically extending out of breadboard base, one horizontally attached via right-angle clamp |
| Optical post right-angle clamp | ThorLabs | RA90 | Connects vertical and horizontal posts |
| Mounting base | ThorLabs | BA1S | Connects optical breadboard base and vertical post |
| 1/4"-20 screw | ThorLabs | SH25S050 | Attaches vertical post to mounting base, mounting base to breadboard base |
| 1/4"-20 set-screw | ThorLabs | SS25E63D | Connects horizontal post to projector via tripod screw-hole |
| Optical power meter | Newport | 840 | Use with power meter detector to measure projector illumination intensity - many power meter models exist, using one that has extendable detector will facilitate measurement |
| Optical power meter detector | Newport | 818-UV | Connects to power meter (above) - UV detector not strictly necessary as blue light is within visible range |
| Adjustable ND filter | K&F Concept | SKU0689 | Adjustable (by rotating) neutral density filter - place above projector aperture |
| Presentation-projector software | Microsoft | Powerpoint | Any software that allows drawing / presentation will suffice |
| CAD software | Autodesk | AutoCAD | Used for designing photomasks, many mask printing services are compatible with AutoCAD files |
| Film photomask | Fineline Imaging | n/a | Many photomask printer services exist for high resolution (>30000DPI) film photomask printing |
References
- Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., Lappin-Scott, H. M. Microbial Biofilms. Annual Review of Microbiology. 49 (1), 711-745 (1995).
- Christensen, B. B., Haagensen, J. A. J., Heydorn, A., Molin, S. Metabolic commensalism and competition in a two-species microbial consortium. Applied and environmental microbiology. 68 (5), 2495-2502 (2002).
- Jin, X., Riedel-Kruse, I. H. Biofilm Lithography enables high-resolution cell patterning via optogenetic adhesin expression. Proceedings of the National Academy of Sciences of the United States of America. 115 (14), 3698-3703 (2018).
- Ohlendorf, R., Vidavski, R. R., Eldar, A., Moffat, K., Möglich, A. From dusk till dawn: one-plasmid systems for light-regulated gene expression. Journal of Molecular Biology. 416 (4), 534-542 (2012).
- Xu, T., et al. Construction of high-density bacterial colony arrays and patterns by the ink-jet method. Biotechnology and Bioengineering. 85 (1), 29-33 (2004).
- Gu, H., Hou, S., Yongyat, C., Detore, S., Ren, D. Patterned Biofilm Formation Reveals A Mechanism for Structural Heterogeneity in Bacterial Biofilms. Langmuir: the ACS Journal of Surfaces and Colloids. 29 (35), 11145-11153 (2013).
- Davey, M. E., O'Toole, G. A. Microbial biofilms: from ecology to molecular genetics. Microbiology and Molecular Biology Reviews: MMBR. 64 (4), 847-867 (2000).
- Nguyen, P. Q., Botyanszki, Z., Tay, P. K. R., Joshi, N. S. Programmable biofilm-based materials from engineered curli nanofibres. Nature Communications. 5, 4945 (2014).
- Chen, A. Y., et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nature Materials. 13, 515-523 (2014).
- JoVE Science Education Database. Basic Methods in Cellular and Molecular Biology. Plasmid Purification. Journal of Visualized Experiments. , (2018).
- JoVE Science Education Database. Basic Methods in Cellular and Molecular Biology. Bacterial Transformation: The Heat Shock Method. Journal of Visualized Experiments. , (2018).
- JoVE Science Education Database. Basic Methods in Cellular and Molecular Biology. Bacterial Transformation: Electroporation. Journal of Visualized Experiments. , (2018).
- JoVE Science Education Database. General Laboratory Techniques. Introduction to Fluorescence Microscopy. Journal of Visualized Experiments. , (2018).
- Pawley, J. . Handbook of Biological Confocal Microscopy. , (2010).
- O'Toole, G. A. Microtiter Dish Biofilm Formation Assay. Journal of Visualized Experiments. 47, e2437 (2011).
- JoVE Science Education Database. Bioengineering. Soft Lithography. Journal of Visualized Experiments. , (2018).
- Levskaya, A., Weiner, O. D., Lim, W. A., Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 461 (7266), 997-1001 (2009).
- Huang, Y., Xia, A., Yang, G., Jin, F. Bioprinting Living Biofilms through Optogenetic Manipulation. ACS Synthetic Biology. 7 (5), 1195-1200 (2018).
- Kac, E. . Signs of Life: Bio Art and Beyond. , (2007).
- Lee, S. A., et al. Trap it!: A Playful Human-Biology Interaction for a Museum Installation. Proceedings of the 33rd Annual ACM Conference on Human Factors in Computing Systems. , 2593-2602 (2015).
- Cira, N. J., et al. A Biotic Game Design Project for Integrated Life Science and Engineering Education. PLOS Biology. 13 (3), e1002110 (2015).
- Bybee, R. W. The next generation science standards and the life sciences. Science and Children. 50 (6), 7 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved