Sign In
Methods Article
Ion Exchange Chromatography (IEX) Coupled to Multi-angle Light Scattering (MALS) for Protein Separation and Characterization
In This Article
Summary
This protocol describes the use of high-specificity ion-exchange chromatography with multi-angle light scattering for an accurate molar mass determination of proteins, protein complexes, and peptides in a heterogeneous sample. This method is valuable for quality assessment, as well as for the characterization of native oligomers, charge variants, and mixed-protein samples.
Abstract
Ion-exchange chromatography with multi-angle light scattering (IEX-MALS) is a powerful method for protein separation and characterization. The combination of the high-specificity separation technique IEX with the accurate molar mass analysis achieved by MALS allows the characterization of heterogeneous protein samples, including mixtures of oligomeric forms or protein populations, even with very similar molar masses. Therefore, IEX-MALS provides an additional level of protein characterization and is complementary to the standard size-exclusion chromatography with multi-angle light scattering (SEC-MALS) technique.
Here we describe a protocol for a basic IEX-MALS experiment and demonstrate this method on bovine serum albumin (BSA). IEX separates BSA to its oligomeric forms allowing a molar mass analysis by MALS of each individual form. Optimization of an IEX-MALS experiment is also presented and demonstrated on BSA, achieving excellent separation between BSA monomers and larger oligomers. IEX-MALS is a valuable technique for protein quality assessment since it provides both fine separation and molar mass determination of multiple protein species that exist in a sample.
Introduction
Quantitative characterization of protein products is increasingly essential as a means of quality control (QC), both for regulatory purposes in the biopharmaceutical industry and to guarantee the reliability and integrity of life science research1,2. As described on the websites of protein networks Protein Production and Purification Partnership in Europe (P4EU) and Association of Resources for Biophysical Research in Europe and Molecular Biophysics in Europe (ARBRE-MOBIEU) (https://p4eu.org/protein-quality-standard-pqs and https://arbre-mobieu.eu/guidelines-on-protein-quality-control, respectively), protein QC must characterize not only the purity of the final product, but also its oligomeric state, homogeneity, identity, conformation, structure, posttranslational modifications, and other properties3,4.
One of the most common QC characterization methods is SEC-MALS. In this method, an analytical SEC column is coupled to MALS and spectrophotometric and refractometric detectors, enabling accurate measurements of the protein molar mass of each peak5. SEC-MALS determines the molar mass of the eluted peaks independently of the elution volume and overcomes the inaccuracy of the analytical SEC using column calibration. The addition of a dynamic light-scattering (DLS) module adds size measurement capability allowing hydrodynamic radius determination. In academic research, SEC-MALS is typically used to determine the oligomeric state of a protein, its conformation, the level of purity, level of aggregation, and modified proteins, such as glycoproteins or lipid-solubilized membrane proteins (determine the molar mass of protein and conjugate the components individually)6,7,8.
In many cases, the final protein of a purification process is not a well-defined molecular species but rather comprises some heterogeneity. The proteins in such a mixture can be varied in terms of structure (for example, different oligomeric forms), conformations, or protein isoforms. Protein heterogeneity can also be a result of minor chemical differences caused by C-terminal lysine processing or asparagine/glutamine deamination, leading to charge variation9,10. Differences in posttranslational modifications such as glycosylation can also lead to heterogeneous samples with charge variations9. These different types of heterogeneity are reflected in the protein’s biophysical characteristics and can impact the stability and biological activity of the target protein11.
Reliable quality control assays of such heterogeneous samples require a highly resolutive analytical separation technique. There are cases where good separation can be challenged to achieve with analytical SEC columns, due to their limited resolution and separation abilities12, resulting in flawed SEC-MALS analysis. Combining a high-specificity separation technique such as IEX with MALS can overcome the limitation of SEC-MALS in heterogeneous samples and provide a complementary method for protein characterization (Table 1 in Amartely et al.12). Unlike SEC, which separates macromolecules by their hydrodynamic size13, IEX separates macromolecules by their surface charge14. Anion exchange (AIEX) and cation exchange (CIEX) matrixes bind negatively and positively charged variants, respectively. With a fine separation between protein populations that share a relatively close mass or shape, IEX-MALS successfully determines the molar mass of each individual protein state in a mixture sample12.
Here we present a standard protocol for running an IEX-MALS experiment for the separation and analysis of BSA oligomeric forms that exist in the same sample. The choice of an IEX column for a specific protein is important and discussed, as well as the pH and conductivity conditions of the buffers. The analysis of IEX-MALS experimental data is also described step by step. Although the separation of BSA oligomers is good and sufficient in SEC-MALS, BSA is a good example to show the IEX-MALS capabilities and to demonstrate the optimization of an experiment. Examples of poor separation achieved by SEC-MALS and proper separation and analysis enabled by IEX-MALS are discussed in a previous study12.
Protocol
1. Preparation of the system
- Install the fast protein liquid chromatography (FPLC) system and MALS/refractive index (RI) detectors (see Table of Materials) along with their respective software packages for control, data acquisition, and analysis per the manufacturers’ instructions.
- Connect the MALS and RI detectors downstream of the FPLC’s UV and conductivity detectors. Bypass the pH detector unless it is absolutely necessary for pH gradients, to minimize the interdetector volume between the UV and MALS detectors. Use capillary tubing of 0.25–0.5 mm inner diameter (i.d.) between the column and detectors and 0.75 mm i.d. capillary tubing on the output of the RI detector to the waste or fraction collector.
- Ensure that the necessary signal connections between the FPLC and detectors have been established, including a UV analog output from the FPLC detector to the MALS Aux input and digital output from the FPLC to the MALS Autoinject In, via the I/O Box.
2. Preparation of the sample and buffer
- Filter all reagents, including the washing and elution buffers, with a 0.1 µm filter. Filter the first 50–100 mL of buffer into a waste bottle in order to eliminate particulates from the dry filters, and keep the remainder of the buffer in a clean, sterile bottle that has been washed thoroughly with filtered water and capped to prevent dust from entering.
- Adjust a BSA sample to a pH and ionic strength (e.g., pH = 8, 50 mM NaCl) that allow binding to the IEX column during dilution, ultrafiltration, or buffer exchange procedures.
NOTE: It is recommended to prefilter the protein sample to the smallest pore size that does not remove the material of interest (0.02–0.1 µm). Alternatively, the sample can be centrifuged at high speed (13,000–16,000 x g) for 10 min to enable the precipitation of large particles. - Prepare at least 0.3–0.5 mg of BSA (see Table of Materials) to inject in a 1 mL column (5/50 mm) for achieving a good-quality MALS analysis. Note that the volume of injection is unlimited.
3. Choice and development of an IEX method for a protein
- Calculate the isoelectric point (pI) of the protein based on the primary sequence, for which servers such as Protparam tool on the ExPASy website can be used15. Note that the pI of BSA is 5.8.
- Select the column type and buffer parameters.
- Use different buffers for AIEX and CIEX, depending on the pKa of the buffer and the ionic nature of the buffer. Use cationic buffers when running the AIEX column and anionic buffers when running the CIEX column with small counterions. In this example, use 20 mM Tris-HCl buffer, pH 8, for the analysis of BSA on an AIEX column.
- For a protein with a pI lower than 7, such as BSA, use an AIEX column and buffer with a pH higher than the pI by at least two units. For a protein with a pI higher than 7, use a CIEX column and buffer with a pH lower than the protein pI by at least two units.
- Optimize the buffer pH according to the strength of binding between the protein and the column matrix. If a protein does not bind well to the column, use a pH farther from the pI. Make sure that the protein is stable at the used pH.
- Use a low salt concentration in the binding and wash the buffers to allow binding of the protein to the matrix, since protein binding depends on the ionic strength of the loaded sample. Proteins usually require some salt for their stability; therefore, use 50 mM NaCl (or alternative salt) during the protein-loading and column-washing steps.
- For elution buffer, use a maximum of 0.5 M NaCl to detach the protein from the column.
NOTE: Higher salt concentrations are not recommended when working with the standard-model RI refractometer due to the range limitation of the instrument. However, a high-concentration model can be used and will accommodate 2 M NaCl.
- Perform an initial method as follows.
- Load 1 mg of BSA (170 µL of 6 mg/mL) in ~0.5 mL of a loading buffer of 20 mM Tris-HCl buffer, pH 8, containing 50 mM NaCl. Wash the column with the same buffer for a 10–15 column volume (CV) to allow the complete elution of the unbound molecules and particles from the system until the light scattering (LS), UV, and RI signals are fully stabilized.
- Perform a short, linear salt gradient of 20–30 CV, using elution buffer of 20 mM Tris-HCl, pH 8, containing 0.5 M NaCl to detach the protein from the column. Perform a gradient of 0%–100% (or alternative widespread gradient) of the elution buffer or split it to two gradients: 0%–50% of elution buffer followed by an additional gradient of 50%–100% of elution buffer, each gradient for 10–20 CV. For BSA, use a widespread gradient of 15%–70% for 30 CV for the initial method.
NOTE: In some instances, the initial method may provide separation for a reliable MALS analysis, and additional runs are not necessary. In many cases, the initial method provides only guidance for further method optimization.
- Optimize the IEX method to increase the resolution and improve the peak separation by changing different parameters.
- Change the gradient slope and length. Short gradients with high slopes provide intense peaks with less separation, while long gradients with mild slopes provide lower peaks with better separation. Find the balance between peak resolution and signal intensity for optimal MALS analysis.
NOTE: Loading a higher quantity of protein can increase LS, UV, and RI signals but does so at the expense of resolution. - Use a stepwise salt concentration profile for the elution. Remember that a combination of steps and linear gradient is commonly used.
- Decrease the flow rate to improve peak separation.
NOTE: For matrixes with very small particles, this is not very significant. - Change the buffer pH to increase the charge variation between the protein populations in the sample and improve the separation between those variants. Note that proteins that are not properly separated at one pH can be separated at a different pH.
- Use a pH gradient, linear or stepwise, to detach proteins from the IEX column. Use elution gradients that combine both pH and salt concentration variations if necessary.
- Use stronger salts, such as MgCl2, or weaker salts, such as sodium acetate, to increase sensitivity and resolution16.
- Use longer IEX columns or a different column matrix with smaller particles to improve separation abilities.
- Change the type of column (AIEX/CIEX) to provide a different pattern of separation. Note that matrixes with strong, weak, or combined (mixed-mode) ligands are also available and can improve the resolution of some samples.
- Use a column from a different supplier with the same ligand that is attached to different matrix resins. The resin itself, independently of the ligand, can interact differently with the proteins and affect the separation profile.
- Include additives (molecules that stabilize the protein in the solution) in the buffers to improve protein stability and avoid protein aggregation, to improve the IEX experiment.
NOTE: Examples of such additives are sugars, alcohols, urea, nonionic or zwitterionic detergents, and chaotropic and kosmotropic salts17,18.
- Change the gradient slope and length. Short gradients with high slopes provide intense peaks with less separation, while long gradients with mild slopes provide lower peaks with better separation. Find the balance between peak resolution and signal intensity for optimal MALS analysis.
4. IEX-MALS experiment
- Open New\Experiment from Method in the MALS software and select the online method from the Light Scattering system methods folder. If the DLS module is available and DLS data are to be acquired, select the online method from the Light Scattering\With QELS subfolder.
- Set the parameters of the run under the Configuration section.
- Set the flow rate of the run in the Generic Pump section to the flow rate used in the FPLC (1.5 mL/min), and enter or verify the buffer parameters under the Solvent section.
- Enter the protein name (BSA), refractive index increment (dn/dc; the standard value for proteins is 0.185 mL/g), UV extinction coefficient at the wavelength of 280 nm (0.66 g/L-1·cm-1), and the concentration of the protein sample (6 mg/mL) in the Sample tab under the Injector section. Insert the sample volume for injection (170 µL) as well under the same section.
- In the Basic Collection tab, under the Procedure section, select the checkbox Trigger on Autoinject and set the duration of the run so that data collection will be continued for at least 5 min after the gradient has reached its final value.
- Set the experiment parameters in the FPLC software.
- Create a new experiment in the Method Editor tab. The initial experiment will be a linear gradient of salt or pH (see step 3.3). For the optimized method, create a more specific gradient or a stepwise program according to the elution results during the initial method (see step 3.4 and Figure 1). Include a pulse signal in the method that will trigger data collection in the MALS software.
- Wash the column and valves with the relevant buffers: 20 mM Tris-HCl, pH 8, with 50 mM NaCl for the washing buffer (A valve) and the same buffer with 500 mM NaCl for the elution buffer (B valve). Make sure that the final column wash uses the binding buffer, containing a low salt concentration, to enable the binding of the protein to the column matrix. For a massive wash of strongly bound impurities, use 0.5 M NaOH before washing with the relevant buffers, followed by a neutralization buffer wash.
- Place the protein sample in the loop using a syringe. If more than 10 mL of the sample is loaded, use a superloop or the pump-valve of the FPLC instrument while bypassing the filter pump and the mixer of the FPLC.
- Start the experiment first in the MALS software by clicking on the Run button and then in the FPLC software. Data will be collected after receiving the pulse signal from the FPLC instrument via the MALS detector.
- Apply the same parameters of the run and instructions described in steps 4.1–4.4 if the IEX-MALS is performed manually with a continuous-flow mode instead of a stand-alone method.
- Once the final method has been verified and run, perform exactly the same method using a blank injection (loading buffer instead of the sample). It is important that the timing between the autoinject pulse and the gradient of the blank run is identical to that of the sample run.
5. Analysis of IEX-MALS experimental data
- Perform the analysis step by step under the Procedure section in the MALS software. The Basic Collection view displays the raw data collection of the experiment.
- Use the Despiking tab to smooth the chromatograms if they exhibit a lot of noise. Normally, use the normal level.
- Define the baseline for all signals (all LS, UV, and RI detectors) in the Baseline view.
- Define the peaks for analysis in the Peaks view. Verify the correct values of dn/dc and the UV extinction coefficient for the protein under each peak.
NOTE: For proteins, it is common to use a standard RI increment value of 0.185 mL/g, but for other macromolecules, different dn/dc values should be used, according to the nature of the molecule. The average dn/dc of polynucleotides (DNA/RNA) is 0.17 mL/g19, while saccharides, such as sucrose, have an average dn/dc value of 0.145 mL/g20 and the dn/dc values of lipids and detergents range between 0.1–0.16 mL/g21. - Analyze the molar mass and the radius using the fitting parameters and the correlation function under the Molar Mass & Radius from LS and Rh from QELS views.
- The RI signal changes significantly during the IEX-MALS run due to the increase in salt concentration. Therefore, subtract the baseline signal from the blank injection for mass calculations that require RI data.
- Open both the protein and blank IEX-MALS experiments. Right-click on the protein experiment name, select Apply Method, and choose the Baseline Subtraction folder from the file dialog. Select the correct type of method (e.g., online) for standard molar mass analysis. Note that the parameters and settings defined for the protein experiment will be saved on the new opened method.
- Under the Baseline Subtraction view, click Import Blank to import the signals of the blank run. Under Instruments (next to the Import Blank button), check all of the detectors to subtract.
- In the Peaks view, adjust the dn/dc values (if necessary) due to the conductivity of the solution at the protein peak area, since the RI of the solution is changed during the run12.
- Calibrate the IEX-MALS system with BSA monomer.
NOTE: Normally, the IEX-MALS system is periodically calibrated for peak alignment, band broadening, and normalization of the angular detectors to the 90° detector using a monodisperse protein with a radius of gyration (Rg) of <10 nm, such as BSA monomer. In this example, BSA serves both as the calibration molecule and is itself the subject of the molar mass analysis.- Align the peaks, under the Procedures > Configuration view.
- Perform normalization under the Normalization view, entering 3.0 nm as the Rg value.
- Under Band Broadening in the same Configuration tab, choose the peak and match the UV and LS signals to the RI signal, using the Perform Fit button.
- The graph of the results is shown in the Results Fitting view. Change the axis scales and other graph parameters by right-clicking on the graph, selecting Edit, and then clicking on the Advanced button. A graph figure with more display options is also available in the EASI Graph tab: select Molar Mass from the Display drop-down menu at the top of the window.
- Note that all results, including molar mass, radius, level of purity, and others, are available in the Report view (summary or detailed) under the Results section. Use the Report Designer button to add more results or parameters, as well as figures, to the report.
Results
BSA is a common protein which is used in chromatography for calibration of the experimental system22 and is highly suitable for practicing IEX-MALS, as well as SEC-MALS. It is primarily monomeric with a theoretical monomer mass of 66.7 kDa and usually incorporates a small number of dimers and higher oligomers23.
BSA was analyzed on IEX-MALS using an anion exchange analytical column (see Table of Materials). A wide linear gradient consisting of 30 CV from 75 mM to 350 mM NaCl separated BSA monomers from the higher oligomers. Downstream MALS analysis resulted in a calculated monomer molar mass of 66.8 ± 0.7 kDa and in a calculated dimer molar mass of 130 ± 5 kDa (Figure 1A).
Based on the buffer conductivity at the eluted peaks, the gradient was changed to a different program: a long step of 175 mM NaCl followed by a linear gradient from 175 mM to 500 mM NaCl. The new gradient greatly improved the resolution and excellent separation between BSA monomer (with a calculated molar mass of 66.1 ± 0.7 kDa) and its higher oligomeric species (with a calculated average mass of 132 ± 2 kDa) (Figure 1B). In order to focus also on the high oligomeric species and to calculate molar masses of each individual oligomeric form of BSA, a stepwise program of 200 mM and 250 mM NaCl was applied. This experiment resulted in an excellent separation between BSA monomer (with a calculated mass of 62.4 ± 0.4 kDa), dimer (with a calculated mass of 130 ± 10 kDa), and trimer (with a calculated mass of 170 ± 10 kDa) (Figure 1C). All IEX-MALS experiments show that BSA elutes mostly as a monomer with a purity of 80%, in agreement with SEC-MALS results where BSA monomers elute with a purity of 85%12.
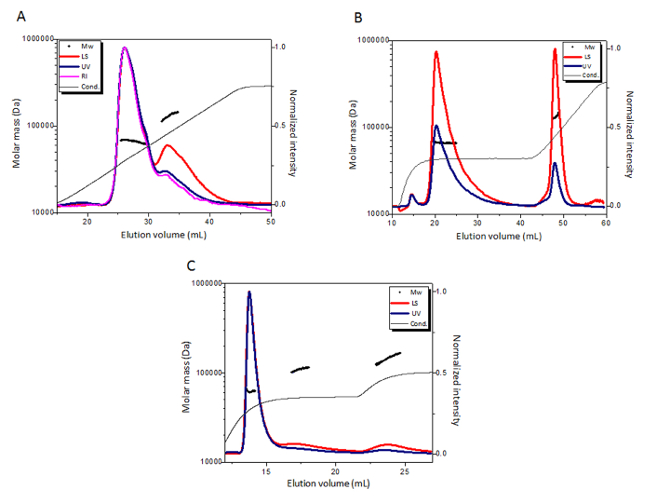
Figure 1: Optimization of IEX-MALS experiment for BSA. (A) IEX-MALS experiment of BSA with a gradient program of 75–350 mM NaCl. (B) IEX-MALS experiment of BSA with a program of a 175 mM NaCl step followed by a linear gradient program of 175–500 mM NaCl. (C) IEX-MALS experiment of BSA with a step program of 200 mM and 250 mM NaCl. The chromatograms display the UV at 280 nm (blue), light scattering at a 90° angle (red), and the refractive index (pink) and the conductivity (grey) curves together with the molar mass of each peak calculated by MALS (black). Please click here to view a larger version of this figure.
Discussion
IEX-MALS is a powerful method for protein separation and characterization that allows the accurate molar mass determination of pure proteins as well as of heterogeneous samples, characterizing native oligomers, nonnative aggregates, covalent and noncovalent complexes, and conjugated proteins. A program consisting of a linear gradient or a series of salt concentration steps can achieve a good separation of the protein populations and allow proper analysis of each individual peak by the MALS. Further optimization by varying different parameters, such as gradient slope (see step 3.4 of the protocol), can be performed if a better resolution is required. IEX-MALS can be a valuable protein quality control assay since it provides an additional, critical level of protein characterization, complementary to other methods such as SEC-MALS.
While SEC-MALS is a standard and common technique for protein molar mass determination, the relatively low resolution of the standard analytical SEC columns may limit accurate molar mass measurements achieved by MALS12. Some examples of the limitations of SEC-MALS include solutions that contain consecutive oligomers, high levels of aggregation that are not fully separated from the monomer peak, and heterogeneous populations with similar molar masses, such as modified proteins.
IEX is a more complex chromatography method to design and carry out than SEC, but the information obtained from an IEX-MALS experiment can complement and sometimes be even more informative than SEC-MALS analysis. IEX-MALS has successfully characterized antibody variants that share the same molar mass, oligomers that are not completely separated on SEC, and short peptides that are difficult to analyze by SEC12,24. Also, macromolecular assemblies that are too large to be separated by SEC, such as full (containing viral DNA) and empty particles of adeno-associated virus (AAV), can be resolved by IEX25 prior to MALS analysis. Compared to SEC, IEX offers more diverse separation capabilities14 and it has the flexibility of adjusting several parameters to increase peak resolution, such as buffer pH, types of salt, types and length of the column, and others. Unlike SEC, there is no volume limitation for sample injection in IEX, and any molecule can be analyzed independent of its size (see Table 1 in Amartely et al.12). This is a great advantage of IEX-MALS, mainly for samples with a low LS intensity, such as very small proteins or diluted proteins with a tendency for aggregation upon concentration. Since analytical IEX columns are more stable and tend to release fewer particles than SEC columns, IEX-MALS requires very short equilibration time, and LS signals are stabilized very fast. This allows the running of individual experiments as described in this protocol and the stopping of the run as required.
Unlike SEC, which usually provides fine results with only one experiment (using a column with the right fractionation range), IEX may require several experiments to achieve optimal resolution by tuning the method parameters. In IEX-MALS experiments that are performed with a salt gradient, the buffer conductivity and, hence, the RI dramatically change during the run, with consequent changes to the RI signal. This requires an additional blank run for each IEX-MALS experiment and an analysis with baseline subtraction (as described in step 5.2 of the protocol) unless the concentration analysis is limited to UV detection (requiring a priori knowledge of the extinction coefficients for each peak). The analysis with baseline subtraction is robust for linear gradients, even though further development of the method is still required for the successful baseline subtraction of salt stepwise programs. The dn/dc values of each peak should be adjusted according to the specific buffer conductivity at the eluted peak (calculations can be found in the literature12). If a protein eluted at an NaCl concentration lower than 200 mM (like in the BSA example), this adjustment is negligible.
The relatively large amount of protein used in IEX-MALS (as detailed in step 2.3 of the protocol) compared to SEC-MALS is important to overcome the dramatic change of the RI signal caused by the salt gradient and the RI fluctuations due to imperfect mixing of the gradient buffers. If only UV detection is used for mass measurement, smaller amounts may be used. The quantity of protein to inject depends on the molar mass of the protein, homogeneity, purity, and the UV extinction coefficient. The necessary injected mass should be higher for smaller proteins and lower for larger proteins (~1 mg for a 20 kDa protein and ~0.2 mg for a 150 kDa protein). Heterogeneous samples require injection of more sample since the quantity is divided between several populations. Analytical high performance liquid chromatography (HPLC) may require less material than an FPLC system.
Recently, in-line analysis using MALS during purification procedures has been reported. Such a real-time analysis is very efficient for detecting aggregation products that occur during purification and can eliminate the need for any further analysis of the protein after purification26. IEX chromatography is frequently used as an intermediate purification step; thus, the combination of preparative IEX columns with MALS can be useful not only as an analytical characterization method but also as a real-time analysis of large-scale purification procedures. Nonanalytical IEX columns are also stable, with a low degree of particle releasing and, therefore, can be used with MALS. Other separation techniques, such as affinity chromatography or hydrophobic exchange chromatography, can also be combined with MALS when subjected to relatively pure samples (to avoid contamination of the MALS and RI detectors). This will require the adaptation and optimization of the method to obtain not only good peak separation but also sufficiently clean LS and RI signals for a successful MALS analysis.
Disclosures
D.S. is an employee of Wyatt Technology Corporation, whose products are utilized in this protocol. A.T. is an employee of Danyel Biotech, a distributor of Wyatt and ÄKTA instruments.
Acknowledgements
The authors thank Dr. Tsafi Danieli (Wolfson Centre for Applied Structural Biology, Hebrew University) for her advice and collaborations. The authors also thank Danyel Biotech Ltd. (Rehovot, Israel) for the assistance and establishment of the analytical FPLC-MALS system utilized in this study.
Materials
| Name | Company | Catalog Number | Comments |
| ÄKTA pure | GE Healthcare | 29-0182-26 | FPLC |
| 0.02 µm Anotop Whatman Filter 10 mm | GE Healthcare | 6809-1002 | Sample Filter |
| 0.1 µm Anotop Whatman Filter 10 mm | GE Healthcare | 6809-1012 | Sample Filter |
| 0.1 µm Anotop Whatman Filter 25 mm | GE Healthcare | 6809-2012 | Mobile phase filter |
| BSA (purity >97%) | Sigma | A1900 | Bovine serum albumin |
| miniDAWN TREOS | Wyatt Technology | WTREOS | MALS |
| mono Q HR 5/50 GL | GE Healthcare | 17-5166-01 | Anion exchange analytical column |
| Optilab T-rEX | Wyatt Technology | WTREX | Refractometer |
| 0.1 mm PES 1000 mL Stericup | Millipore | SCVPU11RE | Mobile phase filter |
| Sodium chloride | Sigma | 71382 | HPLC grade NaCl |
| TRISMA base | Sigma | T-1503 | TRIS buffer |
References
- Lebendiker, M., Danieli, T., de Marco, A. The Trip Adviser guide to the protein science world: a proposal to improve the awareness concerning the quality of recombinant proteins. BMC Research Notes. 7, 585 (2014).
- Raynal, B., Lenormand, P., Baron, B., Hoos, S., England, P. Quality assessment and optimization of purified protein samples: why and how. Microbial Cell Factories. 13, 180 (2014).
- Oliveira, C., Domingues, L. Guidelines to reach high-quality purified recombinant proteins. Applied Microbiology and Biotechnology. 102 (1), 81-92 (2018).
- Manta, B., Obal, G., Ricciardi, A., Pritsch, O., Denicola, A. Tools to evaluate the conformation of protein products. Biotechnology Journal. 6 (6), 731-741 (2011).
- Minton, A. P. Recent applications of light scattering measurement in the biological and biopharmaceutical sciences. Analytical Biochemistry. 501, 4-22 (2016).
- Sahin, E., Roberts, C. J. Size-Exclusion Chromatography with Multi-angle Light Scattering for Elucidating Protein Aggregation Mechanisms. Methods in Molecular Biology. 899, 403-423 (2012).
- Ogawa, T., Hirokawa, N. Multiple analyses of protein dynamics in solution. Biophysical Reviews. 10 (2), 299-306 (2018).
- Rebolj, K., Pahovnik, D., Žagar, E. Characterization of a Protein Conjugate Using an Asymmetrical-Flow Field-Flow Fractionation and a Size-Exclusion Chromatography with Multi-Detection System. Analytical Chemistry. 84 (17), 7374-7383 (2012).
- Liu, H., Gaza-Bulseco, G., Faldu, D., Chumsae, C., Sun, J. Heterogeneity of Monoclonal Antibodies. Journal of Pharmaceutical Sciences. 97 (7), 2426-2447 (2008).
- Hintersteiner, B., et al. Charge heterogeneity: Basic antibody charge variants with increased binding to Fc receptors. mAbs. 8 (8), 1548-1560 (2016).
- Wei, B., Berning, K., Quan, C., Zhang, Y. T. Glycation of antibodies: Modification, methods and potential effects on biological functions. mAbs. 9 (4), 586-594 (2017).
- Amartely, H., Avraham, O., Friedler, A., Livnah, O., Lebendiker, M. Coupling Multi Angle Light Scattering to Ion Exchange chromatography (IEX-MALS) for protein characterization. Scientific Reports. 8 (1), 6907 (2018).
- Goyon, A., et al. Evaluation of size exclusion chromatography columns packed with sub-3μm particles for the analysis of biopharmaceutical proteins. Journal of Chromatography A. 1498, 80-89 (2016).
- Fekete, S., Beck, A., Veuthey, J. -. L., Guillarme, D. Ion-exchange chromatography for the characterization of biopharmaceuticals. Journal of Pharmaceutical and Biomedical Analysis. 113, 43-55 (2015).
- Gasteiger, E., et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Research. 31 (13), 3784-3788 (2003).
- Kopaciewicz, W., Regnier, F. E. Mobile phase selection for the high-performance ion-exchange chromatography of proteins. Analytical Biochemistry. 133 (1), 251-259 (1983).
- Lebendiker, M., Danieli, T. Production of prone-to-aggregate proteins. FEBS Letters. 588 (2), 236-246 (2014).
- Lebendiker, M., Maes, M., Friedler, A. A screening methodology for purifying proteins with aggregation problems. Methods in Molecular Biology. 1258, 261-281 (2015).
- Fu, D., et al. Ultraviolet refractometry using field-based light scattering spectroscopy. Optics Express. 17 (21), 18878-18886 (2009).
- Scafati, A. R., Stornaiuolo, M. R., Novaro, P. Physicochemical and Light Scattering Studies on Ribosome Particles. Biophysical Journal. 11 (4), 370-384 (1971).
- Berthaud, A., Manzi, J., Pérez, J., Mangenot, S. Modeling Detergent Organization around Aquaporin-0 Using Small-Angle X-ray Scattering. Journal of the American Chemical Society. 134 (24), 10080-10088 (2012).
- Umrethia, M., Kett, V. L., Andrews, G. P., Malcolm, R. K., Woolfson, A. D. Selection of an analytical method for evaluating bovine serum albumin concentrations in pharmaceutical polymeric formulations. Journal of Pharmaceutical and Biomedical Analysis. 51 (5), 1175-1179 (2010).
- Hirano, A., Arakawa, T., Kameda, T. Effects of arginine on multimodal anion exchange chromatography. Protein Expression and Purification. 116, 105-112 (2015).
- Avraham, O., Bayer, E. A., Livnah, O. Crystal structure of afifavidin reveals common features of molecular assemblage in the bacterial dimeric avidins. The FEBS Journal. , (2018).
- Lock, M., Alvira, M. R., Wilson, J. M. Analysis of Particle Content of Recombinant Adeno-Associated Virus Serotype 8 Vectors by Ion-Exchange Chromatography. Human Gene Therapy Methods. 23 (1), 56-64 (2012).
- Patel, B. A., et al. Multi-angle light scattering as a process analytical technology measuring real-time molecular weight for downstream process control. mAbs. 10 (7), 945-950 (2018).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
ISSN 2578-2037
Copyright © 2025 MyJoVE Corporation. All rights reserved
We use cookies to enhance your experience on our website.
By continuing to use our website or clicking “Continue”, you are agreeing to accept our cookies.