Method Article
Culture Methods to Determine the Limit of Detection and Survival in Transport Media of Campylobacter Jejuni in Human Fecal Specimens
In This Article
Summary
Although stool culture for Campylobacter is imprecise, it is still considered the gold standard for identification. Methods to determine the limit of detection and survival in transport media of C. jejuni in human stool are described and compared with a new immunoassay with better accuracy.
Abstract
A culture from human stool for diagnosis of Campylobacter-based intestinal illness takes several days, a wait that taxes the fortitude of the physician and the patient. A culture is also prone to false negative results from random loss of viability during specimen handling, overgrowth of other fecal flora, and poor growth of several pathogenic Campylobacter species on traditional media. These problems can confound clinical decisions on patient treatment and have limited the field from answering fundamental questions on Campylobacter growth and infections. We describe a procedure that estimates the lower limit of bacterial numbers that can be detected by a culture and a method for quantifying survival of C. jejuni in media used for transport of this fragile organism. Knowing this information, it becomes possible to set clinically relevant detection thresholds for diagnostic tests and address unstudied issues of whether non-symptomatic colonization is prevalent, if co-infection with other enteric pathogens is common, or if bacterial load correlates with symptoms or serious sequelae. The study also included testing of 1,552 prospectively collected patient diarrheal fecal specimens that were initially classified by conventional culture and further tested by a new enzyme immunoassay. Positive and discrepant specimens were then screened by four molecular methods to assign true-positive or true-negative status. The 5 non-culture methods showed complete agreement on all 48 positive and discrepant specimens, while the culture mis-identified 14 (28%). The specimens that were incorrectly identified by culture included 13 false negative and 1 false positive sample. This basic protocol can be used with multiple Campylobacter spp. and will allow the numbers of Campylobacter bacteria that produce symptoms of gastroenteritis in humans to be determined and for prevalence rates to be updated.
Introduction
The United States Centers for Disease Control (CDC) recently published that the Foodborne Diseases Active Surveillance Network (FoodNet) surveillance program reported 9,723 cases of laboratory-diagnosed Campylobacter infections in 20181. This represents a 12% increase in Campylobacter case reports over 2015−20171. Worldwide, Campylobacter spp. are among the most common bacterial intestinal infections2. Nevertheless, the numbers of Campylobacter-based intestinal illnesses that occur each year are suspected to be underreported3. This underestimation is predictable because most patients can recover with only moderate discomfort and no medical treatment. However, for patients with more severe symptoms or who are at higher risk for serious disease, and who then seek medical care, stool culture is the most common method for assessing whether Campylobacter is the pathogen that is causing their distress4.
For Campylobacter spp., stool culture is particularly troublesome. The most common pathogenic organisms, C. jejuni, C. coli, C. upsaliensis, and C. lari, are microaerophilic5. This means that the bacteria will die at random, unknown rates once exposed to air. The time between specimen collection and culture setup thus becomes an uncontrolled variable in the ability to detect viable Campylobacter spp. by culture.
For direct culture of fecal specimens, the slow growth of Campylobacter is also a problem. Campylobacter colonies are very small even after 48 h of incubation and can easily be covered by competing organisms in the fecal matrix. Plates that contain antibiotics to which most strains of C. jejuni and C. coli are resistant are widely used, as the antibiotics inhibit growth of many (but not all) competing fecal bacteria, allowing better visualization of Campylobacter colonies6. However, other Campylobacter species such as C. lari and C. upsaliensis are sensitive to some of these antibiotics, and either grow poorly or not at all. This contributes to the underreporting of Campylobacter infections from these antibiotic-sensitive species7.
There is a third reason why a culture for Campylobacter may be inaccurate. The bacteria, when stressed, may remain viable but can become "non-culturable"8. This by definition means that the culture will not detect the bacteria present in the sample. How often this occurs is not known8.
Given these potential issues with culture, we used multiple comparison reference methods so that faulty culture results did not make a single comparator assay appear inaccurate9. The culture methods used (e.g., Campylobacter-selective plates, transport medium, gas-generating sachets) were chosen because they are used widely in clinical laboratories for stool specimen culture10.
The culture protocols described here were developed because the lowest number of Campylobacter jejuni that could be detected by culture in human stool was not known. Although estimates have been published for numbers of colony forming units (CFU) present in poultry feces11, these results cannot be equated to human stool, as Campylobacter spp. are commensals in chickens, and do not cause diarrhea. This fundamental information is needed to establish the numbers of Campylobacter bacteria that will produce symptoms of gastroenteritis in humans and to compare virulence between strains or species.
Protocol
1. Enumeration of Campylobacter in Contrived Human Fecal Specimens
NOTE: All steps are carried out using sterile technique and materials on a disposable protective sheet within a disinfected laminar flow safety hood.
CAUTION: Live Campylobacter are infectious and can cause illness, including diarrhea. Wear gloves, a lab coat and safety glasses whenever handling bacteria. Do not mouth pipet. Dispose of all material that has contacted bacteria in proper biohazard containers.
- Growth of stock culture of bacteria
- Obtain strains of C. jejuni (ATCC-33560) or C. coli (ATCC 33559) (Table of Materials) as dried or frozen cultures and rehydrate or thaw bacteria according to manufacturer's instructions. Streak the rehydrated bacteria onto a Campylobacter-specific agar plate to start the culture. Incubate the plate 48 h at 37 °C in an anaerobic jar containing a microaerobic atmosphere gas-generating sachet.
- On the following day, prepare 100 mL of brain-heart infusion (BHI) growth broth containing 0.5% trypticase, 0.5% protease peptone, 0.0125% sodium pyruvate, and 0.0125% sodium bisulfite.
- Prereduce the BHI broth by covering the flask loosely and placing it in an anaerobic jar with a sachet that will produce a microaerophilic environment. Allow broth to prereduce overnight at 37 °C. Similarly, prereduce Campylobacter-specific plates to be used for colony counts in steps 1.1.10 and 1.2.2.
- As Campylobacter are sensitive to air, gather all materials before inoculating broth and do not dawdle while handling cultures. When ready to inoculate, add fetal bovine serum (FBS) to prereduced broth to 4% of total volume. Retain 1 mL of prereduced broth to serve as a blank in measurements of optical density at 600 nm (OD600).
- Remove 3 mL of prereduced broth containing FBS and use broth to scrape the starter plate containing the Campylobacter culture. Gently scrape the plate with an inoculating loop, and then transfer the bacterial slurry to a sterile tube.
- Inoculate the 100 mL of prereduced broth with approximately 3 mL of bacterial slurry and incubate with moderate shaking at 115 rpm at 37 °C in an anaerobic jar containing a gas-generating sachet.
- Monitor growth of the bacteria spectrophotometrically by turbidity at OD600. Use the reserved broth as a blank. If the anaerobic jar is opened, replace the gas-generating sachet.
- Stop broth incubation after 48−72 h or before the OD600 value reaches ~0.4.
NOTE: This OD600 typically equates to 107 to 108 CFU/mL. See Table 1 for typical results. - To establish number of bacteria in pure stock culture, perform eight 10-fold dilution series of 100 µL of broth in 900 µL of dilution buffer (Table of Materials). After the 100 µL of broth has been removed for the first dilution, return flask to anaerobic jar with fresh gas-generating sachet to await use in spiking fecal pool.
- Use sterile plating beads to spread 100 µL of the 10-5 to 10-7 dilutions on duplicate prereduced Campylobacter-specific plates from step 1.1.3. Label plates with dilution used, place them in a second anaerobic jar with gas-generating sachet, and incubate at 37 °C for 48−72 h.
NOTE: See Figure 1 and Figure 2 for dilution scheme and photographs of colonies. - After growth, choose the plate with between 30−300 colonies to count. Utilize the counts to determine the CFU/mL of the stock broth culture using Equation 1:
CFU/mL in stock = Average # of colonies on chosen (duplicate) analytical plates ÷ (mL plated x dilution of plate) [Equation 1]
- Preparation and enumeration of contrived clinical fecal specimens
- Immediately after the plates for analytical counts are prepared in step 1.1.10, make a second set of stock broth dilutions by preparing 10 serial 2-fold dilutions from the stock broth and a Campylobacter-negative fecal pool (NFP). For example, prepare the first dilution by mixing equal volumes of broth and NFP (e.g., 0.1 mL each) and make subsequent dilutions by transferring a designated volume of broth and NFP mixture into a tube with an equal designated volume NFP. Add a control plate with broth containing no Campylobacter added to the fecal pool to help identify non-Campylobacter colonies.
- Make the NFP from de-identified, diarrheal patient surveillance specimens or healthy donor stools that have previously been tested and found to be Campylobacter-negative by methods such as a Campylobacter enzyme immunoassay and by 16S rRNA qPCR.
- T-streak 10 µL of each Campylobacter/stool dilution on duplicate prereduced Campylobacter-specific agar plates. Place plates in the anaerobic jar with a gas-generating sachet and incubate at 37 °C for 48 ± 2 h.
- Examine the streaked plates visually for colonies resembling those from pure Campylobacter cultures.
NOTE: The third quadrant is typically where these will be found. See Figure 1 and Figure 2 for dilution scheme and images of colony size, color and morphology. - Select multiple Campylobacter-like colonies and Gram stain. Using microscopy with an oil immersion lens, examine a thinly streaked area for gram-negative curved, spiral, or cigar-shaped small bacteria.
NOTE: Campylobacter are Gram-negative and require basic fuchsin as a counterstain (instead of the typical safranin) to be visualized accurately. Classic gull-winged bacteria may be seen but are not a requirement. See Figure 2 for representative micrograph. - If either of the duplicate plates at a specific dilution has 1 or more Campylobacter colonies present, consider that dilution fecal-culture positive.
- Consider the last dilution that contains a visible Campylobacter-like gram-negative colony the limit of culture detection. Use Equation 2 to calculate the CFU/mL of the positive dilution in contrived clinical fecal specimen:
CFU/mL in fecal sample = Analytical CFU/mL ÷ Dilution with last positive colony [Equation 2]
NOTE: See Table 2 for typical results.
- Immediately after the plates for analytical counts are prepared in step 1.1.10, make a second set of stock broth dilutions by preparing 10 serial 2-fold dilutions from the stock broth and a Campylobacter-negative fecal pool (NFP). For example, prepare the first dilution by mixing equal volumes of broth and NFP (e.g., 0.1 mL each) and make subsequent dilutions by transferring a designated volume of broth and NFP mixture into a tube with an equal designated volume NFP. Add a control plate with broth containing no Campylobacter added to the fecal pool to help identify non-Campylobacter colonies.
2. Viability Determination of Campylobacter Stored in Transport Media
- Mix 1 mL of Campylobacter broth culture (step 1.1.8) with 1 mL of NFP and prepare 10 duplicate two-fold serial dilutions in NFP. Further dilute each dilution an additional 1:4 in Cary-Blair media, just as a clinical specimen prepared in transport media is treated.
- Store the 20 dilution tubes and a negative control in Cary-Blair medium in capped tubes at 2−8 °C for 96 h and count colonies from each dilution occurring at time zero and every 24 h. For colony counting, sample the broth:fecal tubes and setup fecal culture for colony counting of each dilution, in duplicate.
- Each day plate 10 µL portions of the fecal dilutions on Campylobacter-selective agar and incubate at 37 °C for 48 h, as described above in steps 1.1.9−1.2.7.
- Perform a simultaneous analytical plate count of the original bacterial stock (from step 1.1.8 or a freshly grown broth stock) as described above (steps 1.1.9−1.1.11). Calculate the CFU/mL of the original bacterial stock (Equation 1) to calculate the concentration of bacteria in the transport media fecal sample and its dilutions (Equation 2).
3. Non-culture Assays for Verifying Culture Results
- Use an enzyme immunoassay (EIA) that gives minimal false positive results12 and perform according to package insert instructions to verify culture results.
- Use a molecular assay that can detect the 16S rRNA gene or other gene of a broad range of Campylobacter species13. Confirm that the molecular assay reacts with species such as C. upsaliensis or C. lari that grow poorly on standard antibiotic-containing agar14. Follow manufacturer's instructions for extraction of DNA from fecal samples and performing the test.
NOTE: Bidirectional DNA sequencing of the 16S amplicon can be used to confirm the species of Campylobacter in a positive specimen. Species-specific PCR (see Table 3 for target genes) can also be used to identify species present in discrepant or positive specimens15.
Results
Identifying Campylobacter spp. colonies among competing fecal flora requires keen eyesight and considerable judgement. The lowest number of colonies that can be detected by culture has not been studied, although specimens from patients have been estimated to harbor 106−109 CFU/mL16,17. However, patient samples cannot be used quantitatively as there is no independent method to establish accurate bacterial numbers. To overcome this limitation, two simultaneous measurements are made with one bacterial stock. One test is used for visual detection of Campylobacter colonies from serial dilutions of the stock bacteria in a fecal matrix, simulating clinical specimens; the other is used analytically to quantify the CFU/mL present in the bacterial stock culture used for spiking (Figure 2A).
The detection thresholds for Campylobacter will not be defined values. This is to be expected because each fecal matrix is complex and unique, and growth of bacteria is variable. A key parameter for success is identifying the pinpoint size colonies amongst the competing fecal flora. A representative plate of spiked stool culture is shown in Figure 2C and Figure 2D. The negative control plate without added Campylobacter is important to help identify other fecal flora. Gram staining many candidates also trains the eye to distinguish the correct glossy colonies and the intermediate pink color of fuchsin-stained gram-negative bacteria and confirms the morphology of the bacteria in the selected colonies (Figure 2B). Seven independent experiments were performed, using 5 C. jejuni and 2 C. coli broths, and gave thresholds that overlapped and spanned from 0.3−5 x 106 CFU/mL. See Table 2 for typical data. The detection limits averaged 2 x 106 for C. jejuni and 1.2 x 106 CFU/mL for C. coli. This indicates that culture can likely detect ~1−2 x 106 C. jejuni or C. coli per gram of fecal specimen on standard antibiotic-containing Campylobacter-specific agar used by many clinical laboratories. There are multiple specialized agars with different antibiotics that may give different thresholds for colony detection. The methods described here should encourage more quantitative and comparative studies to improve the accuracy of culture and broaden the versatility of new media. For example, 152 colonies were counted on the first 10-5 plate and 144 colonies on the second 10-5 plate. The average between the two plates is 148 colonies. The plates were inoculated with 0.1 mL (100 µL) of 10-5 dilution, which by Equation 1 equates to 148 x 106 (14.8 x 107) CFU/mL in the pure culture stock. When the fecal dilutions were made, the culture was spiked into negative fecal pool at a 1:1 ratio. Therefore, by Equation 2, the first point (plate "a") on the fecal curve corresponds to 14.8 x 107 divided by 2 and equals 7.4 x 107 CFU/mL. This "a" tube is used to make 9 additional dilutions. In Figure 1, the last dilution with one visible gram-negative colony with Campylobacter-like morphology is on plate "g". This equates to 1.1 x 106 CFU/mL for the fecal culture threshold of detection in this example.
Even though sustained viability is key to culture's accuracy, retention of viability of Campylobacter spp. during handling and shipment of specimens from patients to clinics to reference labs is problematic. Typical storage is to refrigerate specimens in ordinary capped tubes with air exposure and with no special atmosphere. Specimens in transport media (also known as preserved samples) are thought to have better survival, but there are few reports that provide quantitative data18.
The combination of analytical and contrived sample methods shown above was used again to obtain viability and survival time estimates of C. jejuni in transport media. A bacterial stock broth was used to prepare ten duplicate 2-fold to 1024-fold sample dilutions in fecal matrix. The initial broth was found by the analytical counts to have a concentration of 4.8 x 107 CFU/mL. On plates made on day 0, C. jejuni was detected (2 days later) on the plate streaked with the 32-fold dilution, equivalent to 1.5 x 106 CFU/mL. However, on the plates made after refrigerating the Cary Blair fecal sample for 24 hours, only the 2-fold dilution (equivalent to 2.4 x 107 CFU/mL) grew visible colonies. No further loss of viability was seen out to 96 hours, when the study was stopped. This loss of viability equates to a 16-fold (94%) loss of culturable organisms in less than 24 hours and indicates that, even with refrigeration, stool in Cary Blair medium with less than 107 CFU/mL C. jejuni may be missed by culture.
In contrast to the results of culture, the EIA detected the presence of C. jejuni at the 256-fold dilution at the initial time point and throughout the 4 day testing period. The C. jejuni detection threshold for this EIA using spiked fecal samples is 8.4 x 104 CFU/mL. This threshold is below that of fecal culture and allows more sensitive and stable detection of C. jejuni.
To test the ability of culture to detect Campylobacter spp. in an actual clinical setting, 1,552 clinical stool specimens were characterized by 6 procedures: fecal culture, a new immunoassay for Campylobacter spp., and 4 molecular methods. All samples were prospectively collected and initially classified by conventional culture at 3 laboratories in the United States, and then cross-checked by EIA. Any culture-positive or EIA/culture-discrepant specimens were then screened by the molecular methods12. Specimens were assigned a true-positive or true-negative status based on the results of the 5 non-culture methods. The 5 non-culture methods showed complete agreement on all 48 positive and discrepant specimens, while culture mis-identified 14 (28%). The specimens that were incorrectly identified by culture included 13 false negative and 1 false positive sample.

Figure 1: Scheme for simultaneous preparation of analytical and spiked fecal samples. Please click here to view a larger version of this figure.
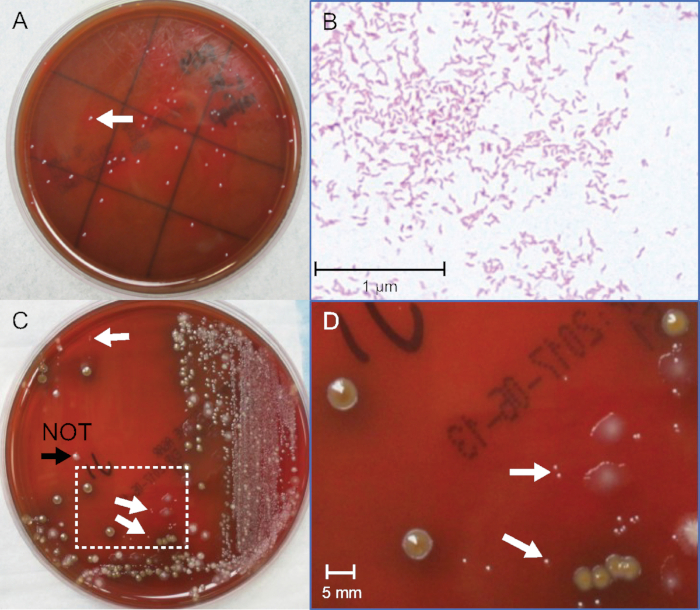
Figure 2: Identification of C. jejuni colonies from pure and fecal cultures. (A) Photograph of C. jejuni colonies from pure bacterial culture after 72 hours incubation. (B) Gram stain of C. jejuni from pure bacterial culture, oil-immersion 400x magnification. (C) Photograph of C. jejuni-positive spiked fecal culture after 48 h incubation. (D) Enlarged area in box in (C), 10x magnification. White arrows indicate pin-point size gram-negative C. jejuni colonies. The black arrowhead indicates a colony that is slightly larger, gram-positive, and not C. jejuni. Please click here to view a larger version of this figure.
| Cultures | OD600 @ T0 | OD600 @ T Final1 | Final CFU/mL |
| C. jejuni | 0.146 | 0.321 | 1.28 x 107 |
| C. coli | 0.245 | 0.508 | 4.50 x 108 |
Table 1: Typical growth and CFU/mL of C. jejuni and C. coli stocks.1 C. jejuni culture was stopped after 48 h of incubation. C. coli culture was stopped after 54 h of incubation.
| Dilution tube for spiked fecal sample | Number of Campylobacter-like colonies | Number of Gram-negative colonies1 | Culture positive? | Calculated CFU/mL of spiked sample | |
| C. jejuni (1.28 x 108 CFU/mL stock) | (2-fold) a | Dense | nd2 | yes | 6.40 x 107 |
| (4-fold) b | 20+ | nd | yes | 3.20 x 107 | |
| (8-fold) c | 4-10 | nd | yes | 1.60 x 107 | |
| (16-fold) d | nd | yes | 8.00 x 106 | ||
| (32-fold) e | nd | yes | 4.00 x 106 | ||
| (64-fold) f | 1-3 | 1 of 2 | yes | 2.00 x 106 | |
| (128-fold) g | 2 of 3 | yes | 1.00 x 106 | ||
| 3(256-fold) h | 1 of 2 | yes | 5.00 x 105 | ||
| (512-fold) i | 0 of 1 | no | 2.50 x 105 | ||
| (1024-fold) j | 0 | nd | no | NFP | |
| C. coli (4.50 x 108 CFU/mL stock) | (2-fold) a | Dense | nd | yes | 2.25 x 108 |
| (4-fold) b | nd | yes | 1.13 x 108 | ||
| (8-fold) c | 50+ | nd | yes | 5.63 x 107 | |
| (16-fold) d | 30+ | nd | yes | 2.81 x 107 | |
| (32-fold) e | 10+ | nd | yes | 1.41 x 107 | |
| (64-fold) f | 3-8 | nd | yes | 7.03 x 106 | |
| (128-fold) g | nd | yes | 3.52 x 106 | ||
| 3(256-fold) h | 1-3 | 1 of 3 | yes | 1.76 x 106 | |
| (512-fold) i | 0 of 1 | no | 8.79 x 105 | ||
| (1024-fold) j | 0 | nd | no | NFP |
Table 2: Typical numbers of colonies on plates of spiked fecal samples.1 Gram negative colonies among Campylobacter-like colonies, 2nd = not determined, 3Data in bold type indicates last positive dilution.
| Species | Gene target |
| C. jejuni | hipO |
| C. coli | cadF |
| C. upsaliensis | cpn60 |
| C. lari | cpn60 |
| C. helveticus | cpn60 |
| C. fetus | cpn60 |
| C. hyointestinalis | cpn60 |
| C. concisus | cpn60 |
Table 3: Genes useful for detection of individual Campylobacter species qPCR.
Discussion
The culture methods described here are built on simple, widely used techniques and materials available in most laboratories10. It is the combination of analytical and contrived samples that provide new information of a clinically relevant detection threshold for fecal cultures. Additionally, the adjudication of culture results with 5 separate assays strengthens the conclusions that Campylobacter fecal culture mis-identifies a significant portion of patient specimens. The EIA and molecular assays are useful as controls because they are each based on a different principle (antigen interaction with antibody vs. DNA amplification) and, importantly, do not rely on viability of bacteria. Note that the EIA assay used for these studies is well-validated and has been shown to agree fully with 4 molecular tests12.
Culture of Campylobacter spp. is particularly troublesome, with sensitivity reported to range from 60−76%19,20, and as evident from its ~30% rate of failure to detect true-positive specimens here. Personnel can expect that control EIA and molecular tests will frequently produce positive results when culture data are negative.
The most critical step in the protocol is the identification of pin-point colonies among competing fecal flora. It is not unusual, as dilutions near the detection threshold, to have alternating zero and non-zero colony count estimates (e.g., 2, 0, 1, 0, 0). It is important to recognize that culture thresholds will be a range of concentrations, not a specific CFU/mL. Nevertheless, the estimate of ~1 x 106 CFU/mL feces as a lower limit for culture detection compares well with reports that infected humans shed 106 to 109 Campylobacter per gram feces21. Changes in antibiotics or agar plates and variations inevitable in individual fecal specimens will undoubtedly change threshold values. This protocol should enable improvements in growth media.
This first information on a limit for culture detection makes it possible to set clinically relevant thresholds for diagnostic tests, and lays the microbiological foundation which is needed to address unstudied issues of non-symptomatic carriage22,23 by Campylobacter, or if bacterial load correlates with symptoms or serious sequelae.
Disclosures
The authors are employees of TECHLAB, Inc. that produces the QUIK CHEK™ kit used as a comparator in this article.
Acknowledgements
These studies were funded by TECHLAB, Inc.
Materials
| Name | Company | Catalog Number | Comments |
| Anaerobic 3.5L Jar | Thermo Fisher | HP0031A | |
| AnaeroGRO Campylobacter Selective Agar | Hardy Diagnostics | AG701 | |
| Bacto Brain Heart Infusion | BD Biosciences | 237500 | |
| Bacto Protease Peptone | Life Technologies Corp | 211684 | |
| Basic Fuchsin | Fisher Scientific | B12544 | |
| BBL Trypticase Peptone | Life Technologies Corp | 211921 | |
| C. coli Type strain | ATCC | 33559 | |
| C. jejuni Type strain | ATCC | 33560 | |
| CampyGen gas generating system sachet | Thermo Fisher | CN0025A | |
| Campylobacter QUIK CHEK | TechLab, Inc. | T5047 / T31025 | |
| Cary-Blair transport medium | Fisher Scientific | 23-005-47 | |
| Coli Roller Sterile plating beads | Millipore Sigma | 71013 | |
| Dilution Buffer | Anaerobe Systems | AS-908 | |
| Fetal bovine serum | Equitech-Bio, Inc | SFBM30 | |
| Sodium bisulfite | Sigma-Aldrich | 243973 | |
| Sodium pyruvate | Sigma-Aldrich | P2256 | |
| Spectrophotometer cuvettes | USA Scientific | 9090-0460 |
References
- CDC. Annual Summaries of Foodborne Outbreaks. , Available from: https://www.cdc.gov/fdoss/annual-reports/index.html (2018).
- Kaakoush, N. O., Castaño-Rodríguez, N., Mitchell, H. M., Man, S. M. Global Epidemiology of Campylobacter Infection. Clinical Microbiology Reviews. 28 (3), 687-720 (2015).
- Pitkanen, T. a, H, M. L. Global Water Pathogen Project. Rose, J. B., Jimenez-Cisneros, B. , UNESCO (2017).
- Fitzgerald, C., et al. Multicenter Evaluation of Clinical Diagnostic Methods for Detection and Isolation of Campylobacter spp. from Stool. Journal of Clinical Microbiology. 54 (5), 1209-1215 (2016).
- Kirkpatrick, B. D., Tribble, D. R. Update on human Campylobacter jejuni infections. Current Opinion in Gastroenterology. 27 (1), 1-7 (2011).
- CDC. Incidence and Trends of Infection with Pathogens Transmitted Commonly Through Food - Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006-2013. Morbidity and Mortality Weekly Report. 63, 328-332 (2014).
- Jaime, A. L., et al. Campylobacter upsaliensis: Another Pathogen for Consideration in the United States. Clinical Infectious Diseases. 34 (11), 59-60 (2002).
- Bullman, S., O'Leary, J., Corcoran, D., Sleator, R., Lucey, B. Molecular-based detection of non-culturable and emerging campylobacteria in patients preseting with gastroenteritis. Epidemiology and Infection. 140, 684-688 (2012).
- Giltner, C. L., Saeki, S., Bobenchik, A. M., Humphries, R. M. Rapid Detection of Campylobacter Antigen by Enzyme Immunoassay Leads to Increased Positivity Rates. Journal of Clinical Microbiology. 51 (2), 618-620 (2013).
- M'ikanatha, N. M., et al. Culturing stool specimens for Campylobacter spp., Pennsylvania, USA. Emerging Infectious Disease. 18, 484-487 (2012).
- Al Amri, A., Senok, A. C., Ismaeel, A. Y., Al-Mahmeed, A. E., Botta, G. A. Multiplex PCR for direct identification of Campylobacter spp. in human and chicken stools. Journal of Medical Microbiology. 56 (10), 1350-1355 (2007).
- Buss, J. E., et al. Campylobacter culture fails to correctly detect Campylobacter in 30% of positive patient stool specimens compared to non-cultural methods. European Journal of Clinical Microbiology & Infectious Diseases. 38, 1087-1093 (2019).
- Wohlwend, N., Tiermann, S., Risch, L., Risch, M., Bodmer, T. Evaluation of a Multiplex Real-Time PCR Assay for Detecting Major Bacterial Enteric Pathogens in Fecal Specimens: Intestinal Inflammation and Bacterial Load Are Correlated in Campylobacter Infections. Journal of Clinical Microbiology. 54 (9), 2262-2266 (2016).
- Couturier, B. A., Hale, D. C., Couturier, M. R. Association of Campylobacter upsaliensis with Persistent Bloody Diarrhea. Journal of Clinical Microbiology. 50 (11), 3792-3794 (2012).
- Chaban, B., Musil, K. M., Himsworth, C. G., Hill, A. K. Development of cpn60-Based Real-Time Quantitative PCR Assays for the Detection of 14 Campylobacter Species and Application to Screening of Canine Fecal Samples. Applied and Environmental Microbiology. 75, 3055-3061 (2009).
- Allos, B., Blaser, M. J. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases, 7th ed. Mandell, G. L., Bennett, J. E., Dolin, R. , Churchill Livingstone. 2793-2802 (2009).
- Anderson, N. W., Buchan, B. W., Ledeboer, N. A. Comparison of the BD MAX Enteric Bacterial Panel to Routine Culture Methods for Detection of Campylobacter, Enterohemorrhagic Escherichia coli (O157), Salmonella, and Shigella Isolates in Preserved Stool Specimens. Journal of Clinical Microbiology. 52 (4), 1222-1224 (2014).
- Wasfy, M., Oyofo, B., Elgindy, A., Churilla, A. Comparison of preservation media for storage of stool samples. Journal of Clinical Microbiology. 33 (8), 2176-2178 (1995).
- Bessède, E., Delcamp, A., Sifre, E., Buissonniere, A., Mégraud, F. New Methods for Detection of Campylobacters in Stool Samples in Comparison to Culture. Journal of Clinical Microbiology. 49 (3), 941-944 (2011).
- Bessède, E., et al. Evaluation of the Diagnostic Accuracy of Two Immunochromatographic Tests Detecting Campylobacter in Stools and Their Role in Campylobacter Infection Diagnosis. Journal of Clinical Microbiology. 56 (4), (2018).
- Shane, A. L., et al. Infectious Diseases Society of America Clinical Practice Guidelines for the Diagnosis and Management of Infectious Diarrhea. Clinical Infectious Diseases. 65 (12), 1963-1973 (2017).
- Toledo, Z., Simaluiza, R. J., Astudillo, X., Fernández, H. Occurrence and antimicrobial susceptibility of thermophilic Campylobacter species isolated from healthy children attending municipal care centers in Southern Ecuador. Revista do Instituto de Medicina Tropical de Sao Paulo. 59, 77-77 (2017).
- Lee, G., et al. Symptomatic and asymptomatic Campylobacter infections associated with reduced growth in Peruvian children. PLoS Neglected Tropical Diseases. 7 (1), 2036(2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved