A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Analysis of Congenital Heart Defects in Mouse Embryos Using Qualitative and Quantitative Histological Methods
In This Article
Summary
In this protocol, we describe procedures to qualitatively and quantitatively analyze developmental phenotypes in mice associated with congenital heart defects.
Abstract
Congenital heart defects (CHD) are the most common type of birth defect in humans, affecting up to 1% of all live births. However, the underlying causes for CHD are still poorly understood. The developing mouse constitutes a valuable model for the study of CHD, because cardiac developmental programs between mice and humans are highly conserved. The protocol describes in detail how to produce mouse embryos of the desired gestational stage, methods to isolate and preserve the heart for downstream processing, quantitative methods to identify common types of CHD by histology (e.g., ventricular septal defects, atrial septal defects, patent ductus arteriosus), and quantitative histomorphometry methods to measure common muscular compaction phenotypes. These methods articulate all the steps involved in sample preparation, collection, and analysis, allowing scientists to correctly and reproducibly measure CHD.
Introduction
CHDs are the most common type of birth defect in humans and are the leading cause of birth defect-related deaths1,2,3,4,5,6. Although about 90% of newborn children survive CHD, it is frequently associated with significant morbidity and medical interventions over the years, imposing a heavy burden on the patients' lives and the healthcare system7,8,9,10. Outside of purely genetic factors, the causes of CHD are poorly understood4. Unidentified causes account for ~56-66% of all CHD cases according to the American Heart Association and other sources2,3,4,11. Well-known factors include genetic mutations, CNVs, de novo single nucleotide variants, and aneuploidy. It is suspected that environmental and dietary factors are also important sources contributing to CHD, as suggested by epidemiological studies linking maternal lifestyle2,12, economic deprivation, and race13, and by research into dietary factors such as folic acid11,14 and the bioactive lipid retinoic acid15,16. Investigating the mechanisms and causes of CHD and other cardiovascular defects is important to develop preventive strategies and novel therapeutic options1,4,17,18,19.
The developing mouse is a cornerstone model for studying CHD in mammals. However, some of the methods and analyses employed, such as dissections preserving heart morphology, analysis of developmental stages, and identification of CHD-associated defects, can be daunting for scientists that are new to the analysis of murine hearts. The goal of the methods described in this protocol is to offer qualitative and quantitative guidelines for these processes. Thus, in this protocol we explain how to perform timed matings to produce embryos of the desired gestational stage, dissect pregnant females for intact heart recovery (including associated tissues such as the outflow tract), heart fixation and preparation for cryostat sectioning, basic histology methods, quantitative analyses of common heart defects, and qualitative analysis of heart muscle compaction, a common precursor phenotype to some types of CHD.
Protocol
All animals used in the experiments referenced in this paper were treated using the animal care guidelines of the Michigan State University Institutional Animal Care and Use Committee (IACUC).
1. Timed mating of C57BL6/J mice for embryo production
- Once mice have reached breeding age (6-8 weeks), put them together in harem breeding format (i.e., two females per one male). Set them up for breeding sometime in the afternoon or evening.
NOTE: Mice often breed within 1 h after lights have been turned off within their housing facility. Females should be retired from breeding at about 6-8 months and males should be used no later than 12 months of age. - If using the weigh-in method to determine fertilization, breed mice in large groups, because the weigh-in method causes timed mating to proceed at a slower pace than methods in which only copulation plugs are monitored.
- The next morning, check for copulation plugs sometime between 8 AM-12 PM. Ensure that this is done at the correct times. If the plug assay is performed earlier than 8 AM, there is a risk that the plug is too deep in the vaginal canal to be observed. If the plug assay is performed later than 12 PM, there is a risk of the plug having fallen out.
- Grab the female by the base of the tail and investigate the opening to the vaginal canal with the use of a micropipette tip. A copulation plug will look like a mucosal barrier (Figure 1A), which might be a bit difficult to see in some cases. Crustiness also is an indication that a copulation plug is or was present. If there is no mucosal barrier (Figure 1B) then the female likely has not mated or the copulation plug could have already fallen out.
- Refer to the morning of copulation as e0.5.
NOTE: With C57BL6/J mice, copulation plugs can be difficult to identify. Therefore, sometimes the mice mated even though no plug was observed. Copulation does not always lead to conception. This is often more likely in C57BL6/J mice. Therefore, monitor weight progression to confirm pregnancy (see step 1.4). The weigh ins can help identify females that are pregnant and confirm that plugs led to conception. Take caution when mating females for consecutive nights. If a particular male is observed to be good at producing a copulation plug, that male can be trusted to mate for consecutive nights with the same female. - Keep in mind that the chances of pregnancy are higher with mice that are proven breeders, as they have successfully been bred before. To increase the chances of having proven breeders, the males used should not be too old.
- Following the plug assay, weigh the females. This weighing should be followed up 7-10 days later. If weight gain exceeds 1.73 g, then the mouse is likely pregnant20. If weight gain is below 1.73 g, it is likely unrelated to pregnancy and the mouse should be reintroduced to the breeding population.
NOTE: C57BL6/J mice produce small litters (five to six embryos on average). Weigh in methods to determine pregnancy are accurate for the majority. Using this method provides a 12.8% false positive rate and will exclude 3% of females that are actually pregnant20. If the researcher is worried that the desired gestational time is later, proceed to step 1.5 - Optional: The day when the dissection is going to take place, ensure that pregnancy has progressed by physical inspection. Grab the female by the base of the tail and stretch the body out. This is easiest to do by placing the mouse on the wrist and gently pulling the base of the tail. If the female is pregnant, then there likely will be weight gain specifically in the lower torso, and it will appear as small lumps.
NOTE: Pregnancy can be palpable as early as e12.5 and will be palpable by e14.5 in most cases. C57BL6/J mice have small litter sizes, therefore the female might be pregnant even if there are no physical signs.
2. Dissection of females and embryos for heart recovery
- Prior to the start of the dissection, prepare the following solutions at room temperature.
- Dissolve ethylenediaminetetraacetic acid (EDTA) into phosphate buffered saline (PBS) at a 0.5 mM concentration.
NOTE: Once prepared, it is preferable to keep the solution on ice and only use it at 4 °C. If contamination is a concern, it should be autoclaved prior to use. - Dissolve paraformaldehyde (PFA) into PBS at a 4% concentration.
NOTE: Once prepared, it is preferable to keep the solution on ice and only use it at 4 °C. Store at 4 °C or colder for long-term storage. The concentration of the PFA solution can vary depending on the downstream applications of the tissue samples. This percentage is used for hematoxylin and eosin staining, immunohistochemistry, and immunofluorescent staining.
- Dissolve ethylenediaminetetraacetic acid (EDTA) into phosphate buffered saline (PBS) at a 0.5 mM concentration.
- Once the female has reached the desired gestational stage, the mouse should be euthanized at the correct time of day according to the approved guidelines of animal use. If timings are on the half day (e.g., e15.5) they should be sacrificed close to 12 PM. If timings are on whole days (e.g., e15.0) they should be sacrificed close to 12 AM.
NOTE: Conception is assumed to occur near midnight during the night the mice are paired. However, it is difficult to determine the exact time of conception. Due to this, timing is estimated, not exact. - After pinning the four limbs down, carefully make an I-shaped incision along the torso, starting from around the urethra up towards the sternum.
- The uterus will likely be located just above the pelvic region. The mouse uterus is Y-shaped; therefore, it will likely be very long and wrapped around the torso. Remove it by gently lifting it. Do this using the sides of fine forceps in a scooping motion. The superficial tissues of the uterus can be grabbed very carefully by the tips of the fine forceps when initially pulling the uterus out. Use scissors to cut the uterus from the body.
NOTE: Keeping in mind the unusual Y-shaped of the uterus, make sure the entire uterus is removed. - Soak the uterus in ice-cold PBS. Pin one end of the uterus down and stretch it out into a single line.
- Cut the uterus into sections, each section containing a separate embryo. Gently peel out the embryo with fine forceps. This is easiest to do with a pair of forceps in each hand. Once removed, place the embryo immediately into a new Petri dish filled with PBS-EDTA solution at 4 °C.
NOTE: Normal PBS solution can also be used, but the EDTA prevents coagulation in the samples. - Stage the embryos using Theiler Staging21. Because staging can vary between embryos within a single litter, stage each individual embryo.
NOTE: This can also provide foresight into the possible congenital heart defects in the embryos. Typically, symptoms of a congenital heart defect can be observed in the embryonic morphology. Other major defects often associated with certain heart conditions can be present. - The heart may be removed in a variety of ways depending on the age of the embryo.
NOTE: If it is unclear whether or not the outflow tract will remain intact during removal, either remove more tissues than solely the heart (keeping the outflow tract intact and minimizing direct contact between the heart and the forceps) or analyze the whole embryo.- Using a dissection scope and two pairs of fine forceps, position the embryo on its back and stabilize access to the chest. This can be done by either pinning the arms down with the forceps or removing the arms and holding the torso in position.
- Use the sharp point of a second pair of fine forceps to make an incision along the sternum of the embryo and extend between the clavicles to the navel. Start by making very fine and superficial incisions while gradually working towards the inside of the chest cavity. The heart should just barely become visible. Do not contact it with the forceps during these superficial incisions.
- Peel the rib cage open using the first pair of forceps to gently squeeze on the torso of the embryo, slightly caudal to the rib cage. The heart should pop out or become apparent. The heart might require some gentle coaxing to come out. Use the side of the forceps, making sure that the point of the forceps never comes in contact with the heart. To keep the workspace and forceps clean, keep a lint-free wipe nearby.
- Gently grab the pulmonary blood vessels with the sides of the forceps, making sure to not sever the tissue, and pull the heart out of the chest cavity. Then, clean the heart.
NOTE: For embryos e13.5 and younger the heart is covered by the pericardium. This must be removed in order to reach the heart.
- Using a stereoscope, take a photograph of the heart for later reference.
- Rinse the hearts in PBS-EDTA solution for 1-2 min, and then place them into 4% PFA solution for approximately 45-60 min to fix them. If analysis of the whole embryo is desired, soak overnight. The volume of the PFA solution should be 5-6x the volume of the tissue being fixed.
NOTE: This incubation time can vary depending on the later studies. This incubation time is used for hematoxylin and eosin staining, immunohistochemistry, and immunofluorescent staining. - Wash 5-10 min with PBS-glycine 3x and place back in PBS. Sodium azide or penicillin/streptomycin can also be added to minimize bacterial growth and preserve intact tissues. The hearts can now be stored short-term at 4 °C.
3. Tissue preparation
NOTE: Tissues can either be prepared using OCT (optimal cutting temperature) embedding or paraffin embedding. There are advantages and disadvantages to either method and the goal of the analysis should be considered when deciding which embedding method should be used.
- OCT embedding
NOTE: This method may be preferable to formalin/paraffin embedding because cryosections preserve antigen reactivity, can still be used for hematoxylin and eosin staining, and produce slices faster.- Prepare a solution of sucrose dissolved into PBS at a 30% (w/v) concentration.
- Transfer the tissues to a new 15 mL conical tube or 1.5 mL microcentrifuge tube containing PBS-30% sucrose. Initially, the tissue will float.
- Place it at 4 °C in the fridge. The tissue will be ready for OCT embedding when it sinks to the bottom of the tube, usually in 24-40 h.
NOTE: Muscle tissues suffer significantly during freeze/thaw cycles and lose their native morphology. Sucrose is absorbed by the tissue and works as a cryopreserving agent that allows improved preservation of the morphology. Make sure there is enough solution in the tube for the floating of the tissues to be sufficiently monitored. PBS-sucrose is easily contaminated with bacteria, so check the solution frequently for cloudiness, which indicates either fungal or bacterial overgrowth. To minimize the risk of contamination either penicillin/streptomycin or sodium azide can be added to the PBS-sucrose solution. - Remove the hearts with a Pasteur pipette, make sure the opening of the pipette is wide enough to not tear the tissue. Cut the pipette with scissors if necessary. If working with whole embryos, use forceps to retrieve the sample.
NOTE: Do this gently. Even if the pipette opening is wide enough to retrieve the hearts, the edges of the pipette can still tear the tissues. Forceps can indent tissue samples if used too aggressively. - Briefly allow the tissue to air dry on a lint-free wipe. Place the sample in an OCT mold in the desired position for cutting (i.e., sagittal, transverse, coronal). Add OCT compound until the sample is completely submerged, making sure there are no bubbles in contact with the tissue.
- Place the mold at -20 °C overnight or several days and then move the mold to -80 °C. After 24 h the mold is ready for cryosectioning. Tissues in this format can be stored long-term.
- Paraffin Embedding
- Using ethanol, dehydrate the tissues in this succession: 50% ethanol for 10 min, 70% ethanol for 10 min, 80% ethanol for 10 min, 95% ethanol for 10 min, 100% ethanol for 10 min (do this 3x).
- Soak the tissues in a series of ethanol/xylene solutions in this succession: 2:1 ethanol/xylene solution for 10-15 min, 1:1 ethanol/xylene solution for 10-15 min, 1:2 ethanol/xylene solution for 10-15 min, 100% xylene for 10-15 min (do this 3x).
- To replace the xylene with paraffin, soak the tissues in a vacuum oven (set between 54-58 °C) in this succession: 2:1 xylene/paraffin solution for 30 min, 1:1 xylene/paraffin solution for 30 min, 1:2 xylene/paraffin solution for 30 min, 100% paraffin for 1-2 h, 100% paraffin for 1-2 h or overnight.
NOTE: Heating the tissues beyond 60 °C for extended periods of time will cause the paraffin polymers to degrade and the tissue to become brittle. - Using fresh paraffin, embed the tissues in the desired position.
4. Cryostat sectioning for OCT embedded tissues
- At the cryostat, place all samples inside for a minimum of 1 h if the samples were previously stored in the -80 °C freezer. They should remain there until all of the sections have been collected.
- Make sure the cryostat is set at the correct temperature settings. The environmental temperature inside the cryostat chamber should be at about -20 °C and the arm of the cryostat should be at about -24 °C.
NOTE: This temperature can vary depending on how the block responds to slicing. If inconsistent slicing is an issue, the temperature of the environment could be lowered. - Remove the OCT block from its mold by pulling at the edges of the open side of the mold to loosen it and then pinching the block at the closed end, where the mold narrows.
- To mount the OCT block, place the room temperature OCT on the chuck, starting from the middle and working to the edge. The entire chuck does not need to be covered.
- Place the OCT block (see step 3.1) on the mold. The edge of the block that was against the closed side of the mold should be facing up on the chuck. Allow the OCT on the chuck to freeze until it is opaque white, about 5-10 min.
- Place the chuck on the arm for another 5-10 min for the block to come to the arm temperature. To get a parallel slice, adjust the arm until the edge of the block is parallel to the stage and there is an even distance between the stage and the bottom edge of the block is the same as the stage and the top edge of the block.
- Insert the blade and adjust the distance of the block from the blade to take sections.
- Take slices at a thickness of 10 µm. Keep slicing until the point of interest is reached either manually or using the trim function.
- To collect slices, use a small flat paintbrush and a detail brush. When the mold begins to be cut, use the flat brush to gently pull the slice against the stand. When cutting through the sample, the movement must be continuous and without pause, although the speed can depend on the firmness of the sample.
- While slicing, use the flat brush to gently guide the slice as it is made. The slice does not need to be flush with the stage at this point, so allow it to billow to not cause tears or pulls and affect histology.
NOTE: When slicing whole embryos, this part can be especially difficult, especially when changing tissue types. Make sure the slices are made with no pauses and the brush does not pull on the slices too aggressively. When tissue types change, the slice can break, so keeping the temperature cold is key. If breaking is an issue, decrease the temperature or increase the thickness of the sample. - Pause the movement of the block once the sample is completely cut through, but the slice is not free from the block yet. Without moving the flat brush from the slice, use the detail brush to pat the slice down to make it flat and freeze against the stage.
- Hold the slice there for ~10 s before removing the detail brush and finishing the slice. Use the two paint brushes to gently flatten the slice against the stage, prevent any rolling, and hold it against the stage for ~20-30 s.
- Flip the slice and flatten against the stage again. Move an electrostatically charged slide close enough for the slice to be attracted and adhere to the slide without having to touch the slide to the stage. Label the slides with a pencil.
- Store the slides in an airtight box to protect the tissues from ice buildup during storage. For short-term storage, slides should be stored at -20 °C before staining. For long-term storage keep them at -80 °C.
5. Microtome sectioning for paraffin embedded tissues
- Place the blocks on ice prior to sectioning in order to chill the samples. When cool, paraffin slices more easily and can produce thinner slices.
- Prepare a 40-45 °C water bath using ultrapure water.
- Put the blade in the holder within the microtome. Ensure it is secure and then set the clearance angle. Make sure the clearance angle is correct. This can be checked following the manufacturer's instructions.
- Place the paraffin block into the microtome. Make sure it is correctly oriented so that the blade will cut straight across the block.
- Ensure the blade is oriented correctly with the block by making a couple of slices. Do this carefully so that adjustments can be made.
- Trim the block until the point of interest is reached.
- Make slices at the desired thickness. Take into account that the first few slices will likely be discarded.
- Pick up the sections and move them into the water bath using forceps. This should make the sections flatten out. Use the forceps to adjust them as needed.
- Move an electrostatically charged slide close enough for the slice to be attracted and adhere to the slide. Label the slides with a pencil.
- Place all collected slides into a slide rack. Allow the slides to dry overnight at 37 °C.
6. Deparaffinization of tissues
- Wash the slides in the following sequence: 3 min in xylene (2x), 3 min in a 1:1 xylene/100% ethanol solution, 3 min in 100% ethanol (2x), 3 min in 95% ethanol, 3 min in 70% ethanol, 3 min in 50% ethanol.
7. Hematoxylin and eosin staining
- Prepare the slides for staining.
- If using OCT prepared tissues, soak in distilled water for ~4 min until OCT is dissolved and let dry.
- If using paraffin embedded tissues, they must be deparaffinized. See step 6.1.
- Place the slide in filtered 0.1% hematoxylin for 10 min. Place the slide in distilled water, changing the water 1x immediately and then again after 3 min.
- Place the slide in 0.5% eosin for 10 s or dip 12x. Dip the slide in distilled water until the eosin no longer streaks, about 2-3 dips.
- Dehydrate the slide by soaking in 50% ethanol for 10 s, 75% ethanol for 10 s, 95% ethanol for 30 s, and 100% ethanol for 1 min. Dip in xylene until xylene comes out smooth, ~5-7 dips.
- Allow xylene to evaporate and mount with a quick drying mounting medium that has a refractive index close to glass.
8. Qualitative analysis of common heart defects
- Take images of all samples using an inverted or stereo microscope and its respective camera. Take macroscopic and microscopic images, depending on the types of defects being analyzed. Macroscopic images can be taken with any magnification below 10x. Microscopic images should be taken at 40x magnification or higher. The photos taken in this paper were taken with 5x magnification using a stereo microscope and a 40x air objective (lens N.A. of 0.55) using an inverted microscope.
NOTE: Macroscopic images are a good starting point because they provide a view to the entire heart (Figure 3). As patterns become identified, specific areas can be focused on and further investigated. - Line up all of the images side by side on a computer screen. Have all of the images of control samples on one side of the screen and all of the images of experimental samples on the other side of the screen. It is best to analyze one treatment group at a time.
- Look for differences in phenotypes between treatment groups, starting at key areas where common heart defects take place. This will vary depending on whether the images are macroscopic, which portray gross anatomy of the heart, or microscopic, which portray microscopic anatomy of the heart tissue.
- Begin searching for common macroscopic heart defects, such as ventricular septal defects (Figure 2A), atrial septal defects (Figure 2B), and patent ductus arteriosus (Figure 2C). All of these defects are most easily seen in the transverse view of the heart.
- To identify septal defects, consider looking at multiple slices, because they might be observed in one plane of the tissue and not another.
NOTE: Sometimes a defect is not the lack of a septum, but a hole in the septum. Therefore, pay careful attention to not mistake a tear for a septal defect. Looking for consistencies between nearby slices in a specific region can confirm if this is actually a defect or a tear from slicing. - To identify defects in the outflow tract, such as the patent ductus arteriosus, use multiple slices to follow the major blood vessels as they leave the heart. Create an image of the major blood vessels relative to one another. An example is featured in Figure 2C.
NOTE: Some irregularities may be due to an embryo's developmental stage (e.g., the patent ductus arteriosus closes postnatally, soon after birth; the ventricular septum does not completely close until ~e14.5). Some other common macroscopic heart defects that are not included in the figures include persistent truncus atereriosus, which can be detected most easily at e16.5 and later; double outlet right ventricle (DORV), which can be detected in slices in which the heart enters the outflow tract; and an overriding aorta22. A common microscopic heart defect includes decreased muscle compaction (Figure 4).
9. Quantitative analysis of cardiac muscle compaction using hematoxylin and eosin stained tissues
- Using the macroscopic view of the stained tissue samples (Figure 4A), identify a region of interest to image at a 40x magnification (Figure 4B). For this paper, the wall of the left ventricle was used. Save the image in the desired format. For this paper, images were saved as .tif files.
- Open the image in ImageJ software.
- Set the image to 8-bit. This allows the image to utilize the threshold tool in the next step23,24. To do this, select "Image | Type | 8-bit".
- Set the threshold of the photo. The purpose of this step is to select only the pixels that represent the background, excluding the pixels that represent the tissues23,24.
NOTE: This surface area will be subtracted later on. A correct threshold will select the pixels that the researcher wants to exclude from muscle tissue surface area. Therefore, the ending result should include the tissue in grayscale and the background will be selected.- Click on "ImageJ | Image | Adjust | Threshold". Move the top bar to make threshold adjustments. Close out the threshold adjustments window without making any other changes once the desired sections are selected21.
- Use the Polygon Selections tool to select the space that the tissue occupies. Carefully make sure the outlines trace the tissue borders, because loose tracings will provide false measurements.
- Adjust the measurement settings on ImageJ so that the software only measures the area of pixels that were selected using the threshold tool in step 9.423. "ImageJ | Analyze | Set Measurements | Area and Limit to Threshold". Select only Area and Limit to Threshold.
- Measure the area of the highlighted pixels. "ImageJ | Analyze | Measure". This value represents the area of the negative space.
- Measure the area of the entire region of interest. This is the area that was selected using the Polygon Selections tool. "Image J | Analyze | Set Measurements | Area". Select only Area and deselect Limit to Threshold. Then measure, selecting "Image J | Analyze | Measure".
- Calculate the area the actual muscle tissue is occupying within the region of interest. Subtract the area of the negative space from the area of the entire region of interest. This value is the area of the muscle tissue.
- Calculate the Muscle Compaction Index (MCI). Divide the area of the muscle tissue by the area of the region of interest.
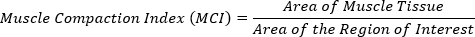
The larger the MCI value, the more compacted the muscle is. These MCI values can then be used for statistical analysis to test for significant changes in muscle compaction between treatment groups (Figure 4C).
Results
The muscle compaction index was compared between hearts developing under two different environments, a control and an experimental group. These protocols were used to analyze the compaction of muscle tissue quantitatively, which permitted statistical analysis. Muscle compaction was shown to be significantly reduced in the experimental hearts relative to the embryos that developed in nonexperimental conditions.
Embryos were disca...
Discussion
This protocol explores the techniques involved in the analysis of cardiac development in embryonic hearts. Some limitations of this method are the required physical dexterity for the preparatory techniques, which may require practice, and skill with microscope imaging. If the slices obtained at the cryostat are messy, the hematoxylin and eosin staining will not be clear, or if the images taken at the microscope have poor lighting, then the method used with ImageJ will not work. A limitation of the threshold feature of Im...
Disclosures
The authors have no disclosures to report.
Acknowledgements
The Aguirre Lab is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under award number K01HL135464 and by the American Heart Association under award number 19IPLOI34660342.
Materials
| Name | Company | Catalog Number | Comments |
| 15 mL Conical Tube(s) | Fisher Scientific | # 1495970C | |
| C57BL/6J Mice | Jackson Labs | C57BL/6J - stock 000664 | |
| Coplin Staining Jars (x6) | VWR Scientific | # 25457-006 | |
| Coverslips 24X50MM #1.5 | VWR Scientific | # 48393-241 | |
| Cryostat - Leica CM3050S | Leica | N/A | |
| Dissecting Dish(s) | Fisher Scientific | # 50930381 | |
| Dumont #5 - Fine Forceps (x2) | Fine Science Tools | # 11254-20 | |
| Eosin Y Solution | Millipore Sigma | # HT110116-500ML | |
| Ethyl Alcohol (Pure, 200 proof) | Fisher Scientific | # BP2818-500 | |
| Ethylenediaminetetraacetic acid (EDTA) | Millipore Sigma | # E9884-100G | |
| Eukitt | Millipore Sigma | # 03989-100ML | |
| Fine Scissors | Fine Science Tools | # 14060-10 | |
| Fluorescent Stereo Microscope Leica M165 FC | Leica | N/A | |
| Glycine | Millipore Sigma | # 410225-250G | |
| Graefe Forceps | Fine Science Tools | # 11052-10 | |
| Graphpad Prism 8 Software | Graphpad | ||
| ImageJ Software | ImageJ | ||
| Kimwipes | Fisher Scientific | # 06666A | |
| Mayer's hematoxylin solution | Millipore Sigma | # MHS16-500ML | |
| Micropipette tip(s) - p200 | Fisher Scientific | # 02707448 | |
| Microsoft Excel Software | Microsoft | ||
| OCT Compound | VWR Scientific | # 102094-106 | |
| Olympus CkX53 Microscope | Olympus | ||
| Paint Brushes (at least 2) | |||
| Paraformaldehyde | VWR Scientific | # 0215014601 | Make into 4% solution (dissolved in PBS) |
| Pasteur pipette(s) | Fisher Scientific | # 13-711-7M | |
| Penicillin-Streptomycin | ThermoFisher Scientific | # 15140122 | |
| Phosphate Buffered Saline (PBS) | ThermoFisher Scientific | # 70011044 | Dilute from 10x to 1x before using |
| Scale | Mettler Toledo | # MS1602TS | |
| Scale | Mettler Toledo | # MS105 | |
| Scalpel Handle #3 | VWR Scientific | # 10161-918 | |
| Scalpel Blades | VWR Scientific | # 21909-612 | |
| Square Mold | VWR Scientific | # 100500-224 | For OCT molds |
| Sucrose | Millipore Sigma | # S9378-500G | |
| Superfrost Plus Slides | Fisher Scientific | # 1255015 | |
| Surgical Scissors | Fine Science Tools | # 14002-14 | |
| Tissue-Tek Accu-Edge Disposable Microtome Blades | VWR Scientific | # 25608-964 | |
| Travel Scale | Acculab | VIC 5101 | |
| Xylene | Millipore Sigma | 214736-1L |
References
- Kathiresan, S., Srivastava, D. Genetics of human cardiovascular disease. Cell. 148 (6), 1242-1257 (2012).
- Sun, R., Liu, M., Lu, L., Zheng, Y., Zhang, P. Congenital Heart Disease: Causes, Diagnosis, Symptoms, and Treatments. Cell Biochemistry and Biophysics. 72 (3), 857-860 (2015).
- Hoffman, J. I. E., Kaplan, S. The incidence of congenital heart disease. Journal of the American College of Cardiology. 39 (12), 1890-1900 (2002).
- Fahed, A. C., Gelb, B. D., Seidman, J. G., Seidman, C. E. Genetics of congenital heart disease: The glass half empty. Circulation Research. 112 (4), 707-720 (2013).
- Pelech, A. N., Broeckel, U. Toward the etiologies of congenital heart diseases. Clinics in Perinatology. 32 (4), 825-844 (2005).
- Zaidi, S., Brueckner, M. Genetics and Genomics of Congenital Heart Disease. Circulation Research. 120 (6), 923-940 (2017).
- Kenny, L. A., et al. Transplantation in the single ventricle population. Annals of Cardiothoracic Surgery. 7 (1), 152-159 (2018).
- Navaratnam, D., et al. Exercise-Induced Systemic Venous Hypertension in the Fontan Circulation. The American Journal of Cardiology. 117 (10), 1667-1671 (2016).
- De Leval, M. R., Deanfield, J. E. Four decades of Fontan palliation. Nature Reviews Cardiology. 7 (9), 520-527 (2010).
- Buckberg, G. D., Hoffman, J. I. E., Coghlan, H. C., Nanda, N. C. Ventricular structure-function relations in health and disease: part II. Clinical considerations. European Journal of Cardio-Thoracic Surgery : Official Journal of the European Association for Cardio-thoracic Surgery. 47 (5), 778-787 (2015).
- Jenkins, K. J., et al. Noninherited risk factors and congenital cardiovascular defects: Current knowledge - A scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young. Circulation. 115 (23), 2995-3014 (2007).
- Botto, L. D., et al. Lower rate of selected congenital heart defects with better maternal diet quality : a population-based study. Archives of Disease in Childhood - Fetal and Neonatal Edition. 101 (1), F43-F49 (2016).
- Knowles, R. L., et al. Ethnic and socioeconomic variation in incidence of congenital heart defects. Archives of Disease in Childhood. 102 (6), 496-502 (2017).
- Feng, Y., et al. Maternal Folic Acid Supplementation and the Risk of Congenital Heart Defects in Offspring : A Meta-Analysis of Epidemiological Observational Studies. Scientific Reports. 17 (5), 8506 (2015).
- Rhinn, M., Dolle, P. Retinoic acid signalling during development. Development. 139 (5), 843-858 (2012).
- Liu, Y., et al. Circulating retinoic acid levels and the development of metabolic syndrome. The Journal of Clinical Endocrinology & Metabolism. 101 (February), 2015 (2016).
- Kurian, L., et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 131 (14), 1278-1290 (2015).
- Aguirre, A., Sancho-Martinez, I., Izpisua Belmonte, J. C. Reprogramming toward heart regeneration: Stem cells and beyond. Cell Stem Cell. 12 (3), 275-284 (2013).
- Srivastava, D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 126 (6), 1037-1048 (2006).
- Heyne, G. W., et al. A simple and reliable method for early pregnancy detection in inbred mice. Journal of the American Association for Laboratory Animal Science. 54 (4), 368-371 (2015).
- . Stage Definition Available from: https://www.emouseatlas.org/emap/ema/theiler_stages/StageDefinition/stagedefinition.html (1998)
- Part 4-Measure Areas using Thresholding. Science Education Resource Center Available from: https://serc.carleton.edu/eet/measure_sat2/part_4.html (2017)
- . Examples of Image Analysis Using ImageJ Available from: https://imagej.nih.gov/ij/docs/pdfs/examples.pdf (2007)
- MacIver, D. H., Adeniran, I., Zhang, H. Left ventricular ejection fraction is determined by both global myocardial strain and wall thickness. IJC Heart and Vasculature. 1 (7), 113-118 (2015).
- Towbin, J. A., Ballweg, J., Johnson, J. Left Ventricular Noncompaction Cardiomyopathy. Heart Failure in the Child and Young Adult: From Bench to Bedside. , 269-290 (2018).
- Choi, Y., Kim, S. M., Lee, S. C., Chang, S. A., Jang, S. Y., Choe, Y. H. Quantification of left ventricular trabeculae using cardiovascular magnetic resonance for the diagnosis of left ventricular non-compaction: evaluation of trabecular volume and refined semi-quantitative criteria. Journal of Cardiovascular Magnetic Resonance. 18 (1), 24 (2016).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved