需要订阅 JoVE 才能查看此. 登录或开始免费试用。
Method Article
药物与DNA相互作用的细胞全基因组映射与COSMIC(小分子交联,染色质隔离)
摘要
识别基因组靶向分子的直接目标仍然是一个重大的挑战。要了解DNA结合分子如何参与基因组,我们开发了依靠小分子交联,染色质隔离的方法(COSMIC)。
摘要
基因组是一些最有效的化学治疗剂的目标,但大多数这些药物缺乏DNA序列特异性,从而导致剂量限制性毒性和许多不良副作用。靶向序列特异性小分子的基因组中可使得分子具有增加的治疗指数和更少的脱靶效应。ñ-methylpyrrole / N甲基咪唑聚酰胺是分子,可以进行合理设计为靶向精致精度特定的DNA序列。不像最自然的转录因子,聚酰胺可以绑定到甲基化和chromatinized DNA无亲和力的损失。聚酰胺的序列特异性已被广泛研究在体外用同源站点识别(CSI),并用传统的生化和生物物理方法,但聚酰胺的细胞结合的基因组的目标的研究仍然难以捉摸。在这里,我们报告的方法,小分子交联伊索拉德染色质(COSMIC),标识在整个基因组聚酰胺结合位点。 COSMIC类似于染色质免疫沉淀,但不同之在两个重要方面:(1)一个光交联剂被用来使选择性的,在时间上控制捕获的聚酰胺结合事件,和(2)的生物素亲和手柄用于纯化聚酰胺半变性条件下-DNA偶联物降低的DNA即非共价结合。宇宙是可用于揭示聚酰胺和其他基因组靶向化学治疗剂的全基因组结合事件一般策略。
引言
的信息,使在人体每个小区被编码在DNA中。有选择地利用这些信息用于调控细胞的命运。转录因子(TF)是结合特定的DNA序列,以表达在基因组中的基因的特定子集蛋白和转录因子的故障是与疾病各种各样的发作,包括发育缺陷,癌症和糖尿病1,2我们一直有兴趣开发分子,可以选择性地结合到基因组中并调节基因调控网络。
聚酰胺由 N个-methylpyrrole的和 N-甲基咪唑能合理设计的分子,可以靶DNA与特异性和亲和力,其竞争对手天然转录因子。3-6这些分子结合在DNA的小沟中的特异性序列。4,5,7 -11聚酰胺已采用两个再加压和激活的特定克的表达埃内斯。4,12-19他们也有有趣的抗病毒20-24和抗癌12,13,25-30性能。聚酰胺之一有吸引力的特点是它们的访问被甲基31,32和缠组蛋白9,10,33 DNA序列的能力。
为了测量DNA结合分子的全面结合特异性,我们实验室建立的同源站点标识符(CSI)的方法。34-39的基于体外的特异性(genomescapes)结合位点的预测的发生可以在基因组上被显示,因为在体外结合强度成正比缔合常数(K a)的 34,35,37,这些genomescapes洞察在整个基因组聚酰胺占用,但测量聚酰胺在活细胞中的结合一直是个难题。 DNA被紧紧包装在细胞核中,它可能会影响结合位点的可接近性。在一个这些chromatinized DNA序列聚酰胺ccessibility仍是一个谜。
最近,许多方法来研究已经出现小分子和核酸之间的相互作用。40-48的化学亲和捕获和大规模并行DNA测序(CHEM-SEQ)是一种这样的技术。的Chem-以次使用甲醛交联的小分子到感兴趣的基因组靶和小分子的感兴趣的生物素化的衍生物捕获配体-靶相互作用。48,49
甲醛的交联导致了可以产生假阳性的间接相互作用。50我们开发了一种新方法,小分子的交联以分离染色质(COSMIC),51与光交联剂,以消除这些所谓的"幻影"的峰。50首先,我们设计并合成聚酰胺的三官能的衍生物。这些分子含有一个DNA结合聚合酶yamide,光交联剂(补骨脂素),和亲和手柄(生物素, 图1)。用三官能聚酰胺,我们可以共价捕获与365nm的紫外线照射下,一个波长不损害DNA或诱导非补骨脂类交联剂聚酰胺-DNA相互作用。51接下来,我们片段的基因组中,净化捕获的DNA在严格的,半-denaturing条件以降低的DNA即非共价结合。因此,我们认为COSMIC作为与化学 - 以次的方法,但用的DNA的更直接读出定位。重要的是,弱(K 10 3 -10 4 M -1)补骨脂用于DNA的亲和力没有可检测的影响的聚酰胺特 异性。51,52富集的DNA片段可以通过定量聚合酶链反应51待分析(COSMIC-qPCR的)或者通过新一代测序53(COSMIC-SEQ)。这些数据使能配体的一个不带偏见,全基因组指导下设计,跨充当与他们所希望的基因位点,并尽量减少脱靶效应。
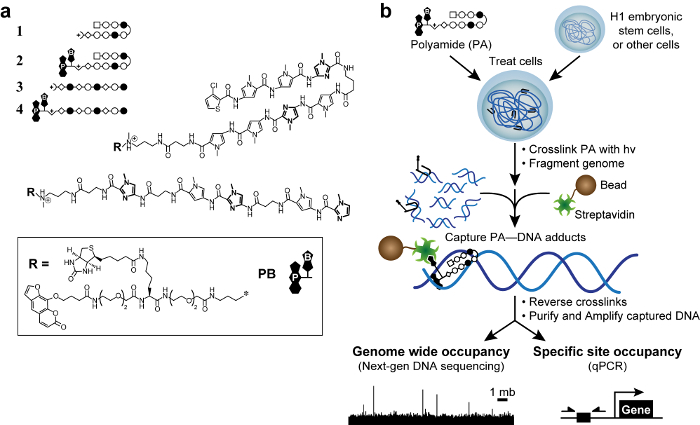
图1.生物活性聚酰胺和宇宙方案 。 (一)夏萍聚酰胺1 - 2靶DNA序列5'-WACGTW-3"。线性聚酰胺3 - 4靶 5'- AAGAAGAAG-3'。 N-甲基咪唑的环以粗体显示的清晰度。开放和充满圆圈表示 N -methylpyrrole 和 N甲基咪唑,分别。方块代表3氯噻吩和菱形代表β丙氨酸。补骨脂素和生物素是由P和B,分别表示。 (二)COSMIC计划。细胞用聚酰胺的三官能的衍生物。用365nm的UV照射交联后,将细胞是升ysed和基因组DNA被剪切。链亲和素包被的磁珠加入到捕获聚酰胺的DNA加合物。该DNA被释放,并可以通过定量PCR(定量PCR)或下一代测序(NGS)进行分析。 请点击此处查看该图的放大版本。
研究方案
1.交联活细胞
- 首先〜2.5×10 7的H1细胞或其它培养的细胞。
注意:此数的H1细胞的对应于5 10-cm的培养皿(约40%铺满)。 - 生长的细胞在E8媒体上碗碟涂覆在支持多能干细胞的表面(参见材料列表),并培育它们在37℃下在5%CO 2的潮湿气氛。酶收获细胞(见材料清单)。注意:不要让H1细胞超过90%汇合;汇合诱导H1细胞的自发分化。要计算细胞,培养细胞的一个额外的10厘米的盘子,抬酶在第1.8-1.10所描述的细胞,并用血球计数。
- 在陈等人准备E8 培养基 。54
- 之前加入聚酰胺,具有连接到真空陷阱巴斯德吸管除去用过的培养基。加入新鲜的介质为erological移液器和分配器(每10厘米的培养皿8ml)中。
注意:介质添加到皿的一侧,以避免破坏细胞。从这点向前保护细胞免受光,以避免过早光交联。 - 添加聚酰胺用吸管(8微升400μM的聚酰胺在DMSO,400nM的终浓度)直接向每个培养皿的培养基。涡流盘均匀地分散在聚酰胺到媒体。
注:浓度可以是多种多样的,但确保没有细胞毒性是观察所选的治疗。 - 孵育细胞24小时,37℃,在5%CO 2的潮湿气氛中,并且确保它们避光。
注:培养时间可以变化,以测量聚酰胺跨越时间绑定。 - 用4ml的PBS(1.05毫磷酸二氢钾,155.17毫氯化钠,2.97 mM磷酸氢二钠),使用血清学移液器和DIS清洗每个10-cm培养皿中penser。吸PBS和中加3毫升E8培养基。
- 用灯光变暗,去掉5培养皿的盖子,然后将细胞在平面上的罩的外侧。放置在5培养皿的玻璃过滤器过滤出的光与λ<300纳米。放置在过滤器的顶部的UV源。交联的样品30分钟,以365纳米的紫外线照射(2.4毫瓦/厘米2)。
注:交联时间应根据经验确定。 - 转移培养皿回罩。与连接到一个真空陷阱巴斯德吸管吸媒体。清洗每个10-cm培养皿用4ml PBS中。吸出PBS。加3毫升酶细胞解离(见材料清单)每10厘米的菜以解离细胞。孵育5分钟,在37℃。
注意 :不要预先暖酶。 - 淬火与每皿3 ml的E8媒体的酶。转移解离细胞向一个15毫升锥形管中。
注:将细胞在冰上从这个点开始。 - CENTRI夫格游离细胞5分钟,500×g离心在4℃下。吸去上清液以除去酶和介质。
注:暂停点 。细胞可以是快速冷冻在液氮中,储存在-80℃下进行后续处理,在以后的时间。
2.隔离染色质
- 添加1.2 ml的COSMIC缓冲液(20mM的Tris-HCl [pH值8.1],2mM EDTA的,150毫摩尔NaCl,1%的Triton-X100,0.1%SDS)中,并移液管上下多次重悬细胞沉淀在每个样品1.2毫升COSMIC缓冲区。
- 加各100mM的苯基甲基磺酰氟(PMSF)133微升,100毫苄脒,和150μM的胃蛋白酶抑制剂蛋白酶抑制剂新鲜至1mM的PMSF和苯甲脒和1.5μM的胃蛋白酶抑制剂的终浓度。细胞裂解物的分裂溶液分成两个琥珀色1.7 ml微量离心管。
- 用超声处理超声仪35分钟(10秒上,10秒关闭,60%的功率),以片段的基因组之间100ND 500基点。
- 保持在平行的微量离心管中染色质溶液的水贮存器中的电平。确认此水平通过视觉检查。
注意:确认DNA的剪切用1.5%琼脂糖凝胶上55 - 优化超声时间经验。使用冰的最小量在储来冷却的样品中,和确保冰不样品和杯角之间的干扰。
- 保持在平行的微量离心管中染色质溶液的水贮存器中的电平。确认此水平通过视觉检查。
- 离心机样品12000×g离心10分钟。保存该水溶液含有水溶性染色质通过将其与吸液管转移到一个新的琥珀离心管中。弃沉淀。
- 转移110微升样品(10%),到一个新的离心管中,贴上标签输入DNA。储存在-80℃。保存该染色质样品的其余部分在冰上在步骤3.3中使用。
3.捕获配体DNA交联的
- 用移液管来分配60微升streptavi在微量离心管嚣包被的磁珠。加入1毫升COSMIC缓冲和组合上的章动5分钟,在室温。放在磁分离磁珠机架2分钟捕捉到珠。除去COSMIC缓冲用吸管。
注意:不要让小球变干。立即进入下一个步骤。 - 添加染色质样品(〜1毫升)珠(60微升),重悬珠子。孵育染色质用链霉亲包被的磁珠至少4小时在旋转,摇动混合机在4℃。如果需要孵育样品O / N。
4.分离亲和纯化的DNA的
- 用7分钟时间在RT用下面的洗涤缓冲液洗珠以除去非特异性相互作用。使用磁分离架上每次洗涤后拍照珠。在洗涤液每次更改后重悬珠。
- 在使用前制备洗涤缓冲液在蒸馏去离子水和过滤器(0.2微米)。商店洗涤缓冲液1和2在4℃下对于several个月。每天准备洗涤液3清新。添加和用吸管除去样品中的洗涤缓冲液。为2和4,加1 和 3(5毫摩),分别在洗涤。样品还可用COSMIC缓冲器下面列出的洗涤(一次12小时,一次4小时)之前,洗涤两次。
- 用洗涤缓冲液1(10mM的三Cl [pH 8.0的],1毫摩尔EDTA,3%SDS)中洗一次。用洗涤缓冲液2(10毫摩尔Tris-氯[pH为8.0],250mM的氯化锂,1mM的EDTA,0.5%NP40,1%脱氧胆酸钠)洗一次。用洗涤缓冲液洗3次(4M尿素,10毫摩尔Tris-氯[pH为7.5],1毫摩尔EDTA,0.1%NP-40)。用Tris EDTA(TE)缓冲液(10mM三Cl [pH 8.0的],1mM EDTA)中洗涤两次。
- 重悬在200μlTE缓冲液珠用吸管。捕获的DNA的该样品被称为亲合纯化的(AP)的DNA。
- 补充输入和AP的DNA具有10交联的逆转缓冲液(100mM的三Cl [pH 8.0的],1M的KOH,4毫EDTA)的1倍终浓度。56,57孵育30分钟,在90℃。
注:254纳米紫外线照射也可以用扭转补骨脂交联,但58环丁基嘧啶二聚体可以从照射而形成在该波长和干扰DNA的下游加工。 - 对于AP的DNA,将装有磁分离样品的离心管架在室温2分钟,分离液体(DNA),用吸管。传送的AP的DNA(其已被释放的珠)到一个新的琥珀离心管中。
注意:所希望的AP的DNA在液体,不再附着于珠。 - 通过用移液管将约1μl的6N HCL中每100微升样品中和的输入和AP的DNA用浓HCl至pH 7。确认样品中加入〜0.5微升至pH试纸用吸管警告中和:将浓HCl是强腐蚀性的酸。根据你的机构处理217; S准则相应的个人防护装备。
- 加入RNA酶A(100毫克/毫升),以0.2微克/微升的最终浓度,以两者的输入和AP的DNA。孵育1小时,在37℃。添加蛋白酶K(20毫克/毫升),以0.2微克/微升的最终浓度,以两者的输入和AP的DNA,加。孵育1小时,在55°C。
- 净化用DNA柱净化试剂盒的DNA(见材料清单)。55洗脱DNA的58微升DNA级H 2 O.存储输入和AP样品在-20℃或-80℃
- 分析美联社DNA的定量聚合酶链反应(qPCR的)与基因座特异性的引物,和/或通过新一代测序。对于定量PCR,采用每个位点2微升AP DNA以下参数:95℃10分钟1个循环和95℃20秒40个循环,54℃或56℃20秒,72℃40秒。
注:退火温度应根据所使用的引物对的熔化温度进行修改。选择一个退火temperatu重,最大限度地减少了定量周期没有非特异性扩增。
注:对于通过新一代测序分析,目的是为至少有1000万映射读取。的映射读取数可以通过增加的AP DNA的量与由较少的样本组合成用于测序运行。更读取提高灵敏度。标准封装的芯片起分析 (例如,荷马,59 MACS,60和最高人民检察院61)与宇宙序列数据进行工作。作为与依靠新一代测序等方法,包括基因组测序和芯片起,重复区域创建对齐和组装含糊不清,因此仍然是一个技术挑战。
结果
为了说明非均匀的基因组片段化和其他变量,纯化的DNA应该总是被规范化对输入DNA的参考。具体到感兴趣的基因座的引物都可以使用。是有帮助的同时分析的轨迹在未预期的分子结合,作为阴性对照。我们看到,在照射> 100倍的聚酰胺占用365纳米的光( 图2)。
富集的DNA也可以通过下一代测序。 DNA被制备用于测序中相同的方式的ChIP的DNA制备(例如,用一...
讨论
One of the primary challenges with conventional ChIP is the identification of suitable antibodies. ChIP depends heavily upon the quality of the antibody, and most commercial antibodies are unacceptable for ChIP. In fact, the Encyclopedia of DNA Elements (ENCODE) consortium found only 20% of commercial antibodies to be suitable for ChIP assays.50 With COSMIC, antibodies are replaced by streptavidin. Because polyamides are functionalized with biotin, streptavidin is used in place of an antibody to capture polyam...
披露声明
A.Z.A. is the sole proprietor of Vista Motif, LLC and WINStep Forward.
致谢
We thank members of the Ansari lab and Prof. Parameswaran Ramanathan for helpful discussions. This work was supported by NIH grants CA133508 and HL099773, the H. I. Romnes faculty fellowship, and the W. M. Keck Medical Research Award to A.Z.A. G.S.E. was supported by a Peterson Fellowship from the Department of Biochemistry and Molecular Biosciences Training Grant NIH T32 GM07215. A.E. was supported by the Morgridge Graduate Fellowship and the Stem Cell and Regenerative Medicine Center Fellowship, and D.B. was supported by the NSEC grant from NSF.
材料
| Name | Company | Catalog Number | Comments |
| Phenylmethylsulfonyl fluoride (PMSF) | any source | ||
| Benzamidine | any source | ||
| Pepstatin | any source | ||
| Proteinase K | any source | ||
| Dynabeads MyOne Streptavidin C1 | Life Technologies | 65001 | |
| PBS, pH 7.4 | Life Technologies | 10010-023 | Other sources can be used |
| StemPro Accutase Cell Dissociation Reagent | Life Technologies | A1110501 | |
| QIAquick PCR Purification Kit | Qiagen | 28104 | We have tried other manufacturers of DNA columns with success. |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 | This Kit can be used to prepare COSMIC DNA for next-generation sequencing |
| Matrigel Basement Membrane Matrix | BD Biosciences | 356231 | Used to coat plates in order to grow H1 ESCs |
| pH paper | any source | ||
| amber microcentrifuge tubes | any source | ||
| microcentrifuge tubes | any source | ||
| pyrex filter | any source | Pyrex baking dishes are suitable | |
| qPCR master mix | any source | ||
| RNase | any source | ||
| HCl (6 N) | any source | ||
| 10-cm tissue culture dishes | any source | ||
| Serological pipettes | any source | ||
| Pasteur pipettes | any source | ||
| Pipette tips | any source | ||
| 15-ml conical tubes | any source | ||
| centrifuge | any source | ||
| microcentrifuge | any source | ||
| nutator | any source | ||
| Magnetic separation rack | any source | ||
| UV source | CalSun | B001BH0A1A | Other UV sources can be used, but crosslinking time must be optimized empirically |
| Misonix Sonicator | Qsonica | S4000 with 431C1 cup horn | Other sonicators can be used, but sonication conditions must be optimized empirically |
| Humidified CO2 incubator | any source | ||
| Biological safety cabinet with vacuum outlet | any source |
参考文献
- Lee, T. I., Young, R. A. Transcriptional Regulation and Its Misregulation in Disease. Cell. 152, 1237-1251 (2013).
- Vaquerizas, J. M., Kummerfeld, S. K., Teichmann, S. A., Luscombe, N. M. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 10, 252-263 (2009).
- Meier, J. L., Yu, A. S., Korf, I., Segal, D. J., Dervan, P. B. Guiding the Design of Synthetic DNA-Binding Molecules with Massively Parallel Sequencing. J. Am. Chem. Soc. 134, 17814-17822 (2012).
- Dervan, P. B. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 9, 2215-2235 (2001).
- Wemmer, D. E., Dervan, P. B. Targeting the minor groove of DNA. Curr. Opin. Struct. Biol. 7, 355-361 (1997).
- Eguchi, A., Lee, G. O., Wan, F., Erwin, G. S., Ansari, A. Z. Controlling gene networks and cell fate with precision-targeted DNA-binding proteins and small-molecule-based genome readers. Biochem. J. 462, 397-413 (2014).
- Mrksich, M., et al. Antiparallel side-by-side dimeric motif for sequence-specific recognition in the minor groove of DNA by the designed peptide 1-methylimidazole-2-carboxamide netropsin. Proc Natl Acad Sci U S A. 89, 7586-7590 (1992).
- Edayathumangalam, R. S., Weyermann, P., Gottesfeld, J. M., Dervan, P. B., Luger, K. Molecular recognition of the nucleosomal "supergroove". Proc Natl Acad Sci U S A. 101, 6864-6869 (2004).
- Suto, R. K., et al. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 326, 371-380 (2003).
- Gottesfeld, J. M., et al. Sequence-specific Recognition of DNA in the Nucleosome by Pyrrole-Imidazole Polyamides. J. Mol. Biol. 309, 615-629 (2001).
- Chenoweth, D. M., Dervan, P. B. Structural Basis for Cyclic Py-Im Polyamide Allosteric Inhibition of Nuclear Receptor Binding. J. Am. Chem. Soc. 132, 14521-14529 (2010).
- Raskatov, J. A., et al. Modulation of NF-κB-dependent gene transcription using programmable DNA minor groove binders. Proc. Natl. Acad. Sci. U.S.A. 109, 1023-1028 (2012).
- Yang, F., et al. Antitumor activity of a pyrrole-imidazole polyamide. Proc. Natl. Acad. Sci. U.S.A. 110, 1863-1868 (2013).
- Mapp, A. K., Ansari, A. Z., Ptashne, M., Dervan, P. B. Activation of gene expression by small molecule transcription factors. Proc. Natl. Acad. Sci. U.S.A. 97, 3930-3935 (2000).
- Ansari, A. Z., Mapp, A. K., Nguyen, D. H., Dervan, P. B., Ptashne, M. Towards a minimal motif for artificial transcriptional activators. Chem. Biol. 8, 583-592 (2001).
- Arora, P. S., Ansari, A. Z., Best, T. P., Ptashne, M., Dervan, P. B. Design of artificial transcriptional activators with rigid poly-L-proline linkers. J. Am. Chem. Soc. 124, 13067-13071 (2002).
- Nickols, N. G., Jacobs, C. S., Farkas, M. E., Dervan, P. B. Modulating Hypoxia-Inducible Transcription by Disrupting the HIF-1–DNA Interface. ACS Chemical Biology. 2, 561-571 (2007).
- Pandian, G. N., et al. A synthetic small molecule for rapid induction of multiple pluripotency genes in mouse embryonic fibroblasts. Sci. Rep. 2, 544 (2012).
- Pandian, G. N., et al. Synthetic Small Molecules for Epigenetic Activation of Pluripotency Genes in Mouse Embryonic Fibroblasts. Chem Bio Chem. 12, 2822-2828 (2011).
- He, G., et al. Binding studies of a large antiviral polyamide to a natural HPV sequence. Biochimie. 102, 83-91 (2014).
- Edwards, T. G., Vidmar, T. J., Koeller, K., Bashkin, J. K., Fisher, C. DNA Damage Repair Genes Controlling Human Papillomavirus (HPV) Episome Levels under Conditions of Stability and Extreme Instability. PLoS ONE. 8, e75406 (2013).
- Edwards, T. G., Helmus, M. J., Koeller, K., Bashkin, J. K., Fisher, C. HPV Episome Stability is Reduced by Aphidicolin and Controlled by DNA Damage Response Pathways. Journal of Virology. , (2013).
- Edwards, T. G., et al. HPV episome levels are potently decreased by pyrrole-imidazole polyamides. Antiviral Res. 91, 177-186 (2011).
- Dickinson, L. A., et al. Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl. Acad. Sci. U.S.A. 95, 12890-12895 (1998).
- Dickinson, L. A., et al. Arresting Cancer Proliferation by Small-Molecule Gene Regulation. Chem. Biol. 11, 1583-1594 (2004).
- Nickols, N. G., et al. Activity of a Py–Im Polyamide Targeted to the Estrogen Response Element. Molecular Cancer Therapeutics. 12, 675-684 (2013).
- Raskatov, J. A., Puckett, J. W., Dervan, P. B. A C-14 labeled Py–Im polyamide localizes to a subcutaneous prostate cancer tumor. Bioorg. Med. Chem. 22, 4371-4375 (2014).
- Jespersen, C., et al. Chromatin structure determines accessibility of a hairpin polyamide–chlorambucil conjugate at histone H4 genes in pancreatic cancer cells. Bioorg. Med. Chem. Lett. 22, 4068-4071 (2012).
- Chou, C. J., et al. Small molecules targeting histone H4 as potential therapeutics for chronic myelogenous leukemia. Molecular Cancer Therapeutics. 7, 769-778 (2008).
- Nickols, N. G., Dervan, P. B. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl. Acad. Sci. U.S.A. 104, 10418-10423 (2007).
- Minoshima, M., Bando, T., Sasaki, S., Fujimoto, J., Sugiyama, H. Pyrrole-imidazole hairpin polyamides with high affinity at 5CGCG3 DNA sequence; influence of cytosine methylation on binding. Nucleic Acids Res. 36, 2889-2894 (2008).
- Warren, C. L., et al. Fabrication of duplex DNA microarrays incorporating methyl-5-cytosine. Lab on a Chip. 12, 376-380 (2012).
- Dudouet, B., et al. Accessibility of nuclear chromatin by DNA binding polyamides. Chem. Biol. 10, 859-867 (2003).
- Carlson, C. D., et al. Specificity landscapes of DNA binding molecules elucidate biological function. Proc. Natl. Acad. Sci. U.S.A. 107, 4544-4549 (2010).
- Warren, C. L., et al. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. U.S.A. 103, 867-872 (2006).
- Tietjen, J. R., Donato, L. J., Bhimisaria, D., Ansari, A. Z., Voigt, C. Chapter One - Sequence-Specificity and Energy Landscapes of DNA-Binding Molecules. Methods Enzymol. 497, 3-30 (2011).
- Puckett, J. W., et al. Quantitative microarray profiling of DNA-binding molecules. J. Am. Chem. Soc. 129, 12310-12319 (2007).
- Keles, S., Warren, C. L., Carlson, C. D., Ansari, A. Z. CSI-Tree: a regression tree approach for modeling binding properties of DNA-binding molecules based on cognate site identification (CSI) data. Nucleic Acids Res. 36, 3171-3184 (2008).
- Hauschild, K. E., Stover, J. S., Boger, D. L., Ansari, A. Z. CSI-FID: High throughput label-free detection of DNA binding molecules. Bioorg. Med. Chem. Lett. 19, 3779-3782 (2009).
- Lee, M., Roldan, M. C., Haskell, M. K., McAdam, S. R., Hartley, J. A. . In vitro Photoinduced Cytotoxicity and DNA Binding Properties of Psoralen and Coumarin Conjugates of Netropsin Analogs: DNA Sequence-Directed Alkylation and Cross-Link. 37, 1208-1213 (1994).
- Wurtz, N. R., Dervan, P. B. Sequence specific alkylation of DNA by hairpin pyrrole–imidazole polyamide conjugates. Chem. Biol. 7, 153-161 (2000).
- Tung, S. -. Y., Hong, J. -. Y., Walz, T., Moazed, D., Liou, G. -. G. Chromatin affinity-precipitation using a small metabolic molecule: its application to analysis of O-acetyl-ADP-ribose. Cell. Mol. Life Sci. 69, 641-650 (2012).
- Rodriguez, R., Miller, K. M. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nat Rev Genet. 15, 783-796 (2014).
- Guan, L., Disney, M. D. Covalent Small-Molecule–RNA Complex Formation Enables Cellular Profiling of Small-Molecule–RNA Interactions. Angew. Chem. Int. Ed. 52, 10010-10013 (2013).
- White, J. D., et al. Picazoplatin, an Azide-Containing Platinum(II) Derivative for Target Analysis by Click Chemistry. J. Am. Chem. Soc. 135, 11680-11683 (2013).
- Rodriguez, R., et al. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 8, 301-310 (2012).
- Bando, T., Sugiyama, H. Synthesis and Biological Properties of Sequence-Specific DNA-Alkylating Pyrrole−Imidazole Polyamides. Acc. Chem. Res. 39, 935-944 (2006).
- Anders, L., et al. Genome-wide localization of small molecules. Nat. Biotechnol. 32, 92-96 (2014).
- Jin, C., et al. Chem-seq permits identification of genomic targets of drugs against androgen receptor regulation selected by functional phenotypic screens. Proc. Natl. Acad. Sci. U.S.A. 111, 9235-9240 (2014).
- Landt, S. G., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research. 22, 1813-1831 (2012).
- Erwin, G. S., Bhimsaria, D., Eguchi, A., Ansari, A. Z. Mapping Polyamide–DNA Interactions in Human Cells Reveals a New Design Strategy for Effective Targeting of Genomic Sites. Angew. Chem. Int. Ed. 53, 10124-10128 (2014).
- Hyde, J. E., Hearst, J. E. Binding of psoralen derivatives to DNA and chromatin: influence of the ionic environment on dark binding and photoreactivity. Biochemistry. 17, 1251-1257 (1978).
- Erwin, G. S., Bhimsaria, D., Rodríguez-Martínez, J. A., Grieshop, M. P., Ansari, A. Z. Genome-wide localization of polyamide-based genome readers reveals sequence-based binding to repressive heterochromatin. In preparation. , (2015).
- Chen, G., et al. Chemically defined conditions for human iPSC derivation and culture. Nat Meth. 8, 424-429 (2011).
- Deliard, S., Zhao, J., Xia, Q., Grant, S. F. A. Generation of High Quality Chromatin Immunoprecipitation DNA Template for High-throughput Sequencing (ChIP-seq). J Vis Exp. (74), e50286 (2013).
- Shi, Y. B., Spielmann, H. P., Hearst, J. E. Base-catalyzed reversal of a psoralen-DNA cross-link. Biochemistry. 27, 5174-5178 (1988).
- Kumaresan, K. R., Hang, B., Lambert, M. W. Human Endonucleolytic Incision of DNA 3′ and 5′ to a Site-directed Psoralen Monoadduct and Interstrand. J. Biol. Chem. 270, 30709-30716 (1995).
- Cimino, G. D., Shi, Y. B., Hearst, J. E. Wavelength dependence for the photoreversal of a psoralen-DNA crosslink. Biochemistry. 25, 3013-3020 (1986).
- Heinz, S., et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell. 38, 576-589 (2010).
- Zhang, Y., et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
- Kharchenko, P. V., Tolstorukov, M. Y., Park, P. J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotech. 26, 1351-1359 (2008).
- Diamandis, E. P., Christopoulos, T. K. The biotin-(strept)avidin system: principles and applications in biotechnology. Clin. Chem. 37, 625-636 (1991).
- Martinson, H. G., True, R. J. On the mechanism of nucleosome unfolding. Biochemistry. 18, 1089-1094 (1979).
- Gloss, L. M., Placek, B. J. The Effect of Salts on the Stability of the H2A−H2B Histone Dimer. Biochemistry. 41, 14951-14959 (2002).
- Jackson, V. Formaldehyde Cross-Linking for Studying Nucleosomal Dynamics. Methods. 17, 125-139 (1999).
- Kasinathan, S., Orsi, G. A., Zentner, G. E., Ahmad, K., Henikoff, S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Meth. 11, 203-209 (2014).
- Teytelman, L., Thurtle, D. M., Rine, J., van Oudenaarden, A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 18602-18607 (2013).
- . Phantompeakqualtools home page Available from: https://www.encodeproject.org/software/phantompeakqualtools/ (2010)
- Wang, D., Lippard, S. J. Cellular processing of platinum anticancer drugs. Nature Reviews Drug Discovery. 4, 307-320 (2005).
- Hurley, L. H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2, 188-200 (2002).
转载和许可
请求许可使用此 JoVE 文章的文本或图形
请求许可探索更多文章
This article has been published
Video Coming Soon
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。