需要订阅 JoVE 才能查看此. 登录或开始免费试用。
Method Article
中子自旋回声光谱学作为脂膜动力学和膜-蛋白相互作用的独特探针
Erratum Notice
摘要
本文介绍了中子自旋回声(NSE)脂质膜研究中的样品制备、数据减少和数据分析方案。脂质的明智标记使获得中等长度和时间尺度上的不同膜动力学,从而发生重要的生物过程。
摘要
脂质双层体是细胞膜的主要基质,是营养交换、蛋白膜相互作用和病毒萌芽等重要细胞过程的主要平台。对于有效的生物活性,细胞膜应足够刚性,以保持细胞及其隔间的完整性,但流动性足以使膜成分(如蛋白质和功能领域)扩散和相互作用。这种弹性和流体膜特性的微妙平衡及其对生物功能的影响,需要更好地了解关键生物过程(例如膜变形和蛋白质结合事件)的中观长度和时间尺度上的集体膜动力学。能够有效探测这一动态范围的技术包括中子自旋回波(NSE)光谱学。结合二元标签,NSE可用于直接访问弯曲和厚度波动以及选择膜特征的中等动力学。本文简要介绍了NSE技术,概述了在脂膜上进行NSE实验的程序,包括样品制备和去子宫化方案的细节,以及数据收集和减少说明。本文还介绍了用于提取关键膜参数的数据分析方法,如弯曲刚性模态、区域可压缩模态和平面粘度。为了说明NSE研究的生物学重要性,探讨了NSE所探讨的膜现象的选例,即添加剂对膜弯曲刚性的影响、域形成对膜波动的影响以及膜蛋白相互作用的动态特征。
引言
在过去的几十年里,对细胞膜及其功能的理解有了显著的演变。以前认为细胞膜是定义细胞边界和家膜蛋白1的被动脂质双层蛋白,现在已逐渐转变为一种动态模型,其中脂质双层层在调节重要的生物过程(包括细胞信号、分子交换和蛋白质功能)方面发挥着重要作用,仅举几例2、3、4、5、6。认识到细胞膜具有高度动态性,不断进行改造和分子再分配,促使科学探索膜7、8、9的平衡结构。因此,已经开发了多种方法来研究生物和生物吸血脂膜中的各种动态模式。迄今为止,这些研究大多集中在扩散分子运动10,11,12,13和宏观形状波动14,15,16,留下一个显着的差距,了解中间膜动力学,即脂质组合体的集体波动,包括很少10-100的脂质分子。这些动态发生在几十到几100+的长度尺度上,以及随着时间尺度的子 n 到几百 ns(见图 1),这里称为中等尺度。事实上,正是在这些尺度上,关键的生物活动发生在膜17层。这包括病毒萌芽18,通道盖19,膜蛋白相互作用20。同样重要的是要指出,膜蛋白的能量景观21,22表明,蛋白质的构象变化-必要的调节作用-发生在23的集体膜波动的ns时间尺度,进一步强调间皮动力学在细胞膜的生物功能及其生物灵感类比20的重要性。本文重点研究脂质膜中两种主要的中观动态模式,即弯曲波动和厚度波动。
直接探测这些波动模式的主要挑战是难以同时使用标准光谱法访问其空间和时间尺度。另一个挑战是,直接接触技术可能会影响相同的波动,他们打算测量16。生物膜24、25的组成和结构复杂性进一步加剧了这种情况,导致非同质膜特征,包括脂质域形成26、27、28、29、30和膜不对称31、32、33要求选择性探针来了解不同膜特征的动态。幸运的是,这些挑战可以通过非侵入性中子光谱方法克服,如中子自旋回波(NSE),它固有地访问所需的长度和时间尺度,并进一步使选择性膜特征的研究不改变其物理化学环境34。事实上,在过去几年中,NSE光谱学已经发展成为一个独特而强大的集体膜动力学探测器35。NSE对脂质膜的研究结果对脂膜的机械36、37和粘性38、39特性有了新的认识,并揭示了脂膜在生物功能40、41中的潜在作用。
NSE光谱技术基于干涉测量仪器设计,最初由Mezei42提出,使用一系列自旋翻转器和磁线圈来控制中子自旋的预兆,因为中子穿过仪器。设计基于与样品位置有关磁场元素的磁反射(图1A)。这意味着,在中子和样品之间没有能量交换的情况下,中子在仪器的前半部分和后半部分(注意两个预切线圈之间的π翻转器)以相反的方向执行相同数量的自旋预演。"因此,中子的最终自旋状态相对于初始状态保持不变 - 一种称为自旋回声的现象(见图1A中的透明中子)。然而,当中子与样品进行能量相互作用时,能量交换会改变仪器后半部分的自旋前额,从而导致不同的最终自旋状态(见图1A)。这是实验性地检测为极化损失,稍后将在本文中显示。有关 NSE 技术的更多详细信息,读者将参考专用技术论文 42、43、44、45。
在此处,提供了简化的描述,以提供 NSE 可访问的长度和时间尺度的粗略估计。长度刻度由可实现的波维克转移范围(Q = 4 = sin = /é)决定,其中2 = 是散射角,+是中子波长。可以看到Q是由光谱仪第二臂的波长范围和旋转范围设置的(见图 1A)。NSE 光谱仪上典型的Q范围为 +0.02-2 =-146、47和高达 0.01-4 +-1,最近升级为 48,49,对应于 1-600 + 的空间尺度。另一方面,无障碍时间尺度是根据磁预切线圈内中子获得的总预切角(或相位)计算得出的,并且发现为50: 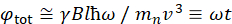 。在这个表达中,t是傅立中的时间定义为
。在这个表达中,t是傅立中的时间定义为 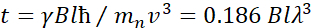 ,
, 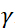 中子陀螺仪磁比在哪里,
中子陀螺仪磁比在哪里,  是线圈长度,
是线圈长度, 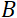 是线圈磁场的强度。值得指出的是,傅立叶时间是严格依赖于仪器几何、磁场强度和中子波长的数量。例如,使用波长 = 8 + 的中子
是线圈磁场的强度。值得指出的是,傅立叶时间是严格依赖于仪器几何、磁场强度和中子波长的数量。例如,使用波长 = 8 + 的中子 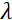 和
和  1.2 米和
1.2 米和 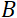 + 0.4 T 的仪器设置,Fourier 时间计算为t = 50 ns。在实验中,Fourier 时间通过改变预切线圈中的电流(即磁场强度)或使用不同的中子波长进行调整,从而产生典型的 NSE 时间尺度 =1 ps 到 100 ns。但是,最近 NSE 光谱仪的升级使访问时间更长的 Fourier 时间, 海因茨迈尔-莱布尼茨赞特鲁姆51号的 J-NSE-Phoenix 光谱仪和橡树岭国家实验室48号的 SNS-NSE 光谱仪高达 400 ns,以及劳兰格文学院 (ILL) 49 的 IN15 NSE 光谱仪上高达1,000ns。
+ 0.4 T 的仪器设置,Fourier 时间计算为t = 50 ns。在实验中,Fourier 时间通过改变预切线圈中的电流(即磁场强度)或使用不同的中子波长进行调整,从而产生典型的 NSE 时间尺度 =1 ps 到 100 ns。但是,最近 NSE 光谱仪的升级使访问时间更长的 Fourier 时间, 海因茨迈尔-莱布尼茨赞特鲁姆51号的 J-NSE-Phoenix 光谱仪和橡树岭国家实验室48号的 SNS-NSE 光谱仪高达 400 ns,以及劳兰格文学院 (ILL) 49 的 IN15 NSE 光谱仪上高达1,000ns。
除了直接访问膜动力学的长度和时间尺度外,NSE还具有中子同位素灵敏度的固有能力。具体来说,中子与氢同位素(生物系统中最丰富的元素)的相互作用能力,导致不同的中子散射长度密度,34或NSLD(相当于折射50的光学指数),当正二氧化硅被钚取代时。这支持一种称为对比度变异的方法,通常用于突出特定膜特征或隐藏其他特征-后一种方案称为对比匹配。对比变异/匹配的频繁应用是用水(NSLD = -0.56 × 10-6 +-2)用重水或D2O(NSLD = 6)代替水 .4 × 10-6 +-2) 以放大来自亲生脂质膜的中子信号 (NSLD = 0 × 10-6 +-2)。这种方法在膜结构研究中非常有效,因为D2O渗透到膜的头部群区域,可以准确确定膜厚度(见图2A,左面板)和不同脂质子组的位置,当应用更复杂的模型53,54。本文重点介绍了生物仿生膜中集体动力学研究的对比变异和选择膜特征的一些例子。
在这里,NSE 通过 NSE 对模型和生物相关脂质膜系统进行有形研究的有形示例,以脂质悬浮的形式,强调独立膜中的中尺度动力学,从而说明了 NSE 在提供对动态和功能膜特性的独特见解方面的有效性。对于NSE对平面膜动力学的测量,读者指的是关于放牧发生中子自旋回波光谱(GINSES)55,56和其他对齐多拉梅拉膜堆栈57,58,59,60的研究的专门出版物。
为了简单起眼,本文重点介绍了三种不同的膜去质方案,这些方案在经过充分研究的域形成中得到了说明, 或相分离,脂质双层系统1,2-二甲基-sn-甘油-3磷胆碱(DMPC)和1,2-脱脂-sn-甘油-3-磷胆碱(DSPC)混合物61,62。这两种脂质的特点是碳氢化合物链长度不匹配(DMPC 中的 14 碳/尾与 DSPC 中的 18 碳/尾)及其凝胶流体过渡温度(Tm、DMPC = 23 °C vs Tm、DSPC = 55 °C)。这导致在DMPC:DSPC膜的横向相分离在混合物63的上下过渡温度之间的温度。此处考虑的去质方案旨在演示 NSE 测量中对脂质膜的不同动态模式,即弯曲波动、厚度波动和横向域的选择性弯曲/厚度波动。所有脂质成分均报告为 DMPC:DSPC 双层,以 70:30 的摩尔分数为小数,使用 DMPC 和 DSPC 的商用亲生和渗透变体。所有样品制备步骤均基于 D2O 中的 4 mL 脂质悬浮液,脂质浓度为 50 毫克/mL,每个样品的总脂质为Mtot = 200 mg。
研究方案
1. 实验所需的解育方案
- 对于弯曲波动测量,在 D 2 O (D99.9%)或 D2O 缓冲(例如,用 D 2 O 而不是H2O 制备磷酸盐缓冲器)中制作完全亲生脂质体。使用完全增殖的 DMPC (C36H72NO8P) 和 DSPC (C44H88NO8P) 与
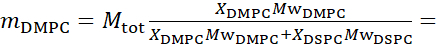 133.4 毫克, 其中XDMPC和XDSPC是 DMPC 和 DSPC 的摩尔分数,此处分别设置为 0.7 和 0.3,MwDMPC和 MwDSPC分别为 677.9 克/摩尔和 790.1 克/摩尔给出的摩尔重量。同样,mDSPC = 66.6 毫克。此去位方案增加了膜 (NSLD = 0 × 10-6 +-2)和去位缓冲区 (NSLD = 6.4 × 10-6 +-2)之间的散射对比,并放大了膜起伏的信号(见图 2A左面板)。
133.4 毫克, 其中XDMPC和XDSPC是 DMPC 和 DSPC 的摩尔分数,此处分别设置为 0.7 和 0.3,MwDMPC和 MwDSPC分别为 677.9 克/摩尔和 790.1 克/摩尔给出的摩尔重量。同样,mDSPC = 66.6 毫克。此去位方案增加了膜 (NSLD = 0 × 10-6 +-2)和去位缓冲区 (NSLD = 6.4 × 10-6 +-2)之间的散射对比,并放大了膜起伏的信号(见图 2A左面板)。 - 用于测量选定横向膜特征的弯曲动力学,例如相分离 DMPC:DSPC 膜中的矩阵动力学, 使用催化 DMPC (C36H72NO8P) 和除臭, DSPC-d83 (C44H5NO8PD83,Mw 873.7 g/mol), 使 mDMPC = 128.8 毫克和 mDSPC-d83 = 71.2 毫克.此去位化方案最大限度地减少了来自不受欢迎的 DSPC 富润域的散射,从而能够选择性地测量 DMPC 富含矩阵的弯曲波动(见 图 2B 中间)。
注:为了找到特定对比匹配方案所需的最佳脂质去质,请使用可用的基于 Web 的散射长度密度 (SLD) 计算器,例如 NIST 中子研究中心64开发的计算器。这些基于 Web 的界面配备了用户友好的工具,便于计算不同程度的脱脂脂质的 SLD 以及脂质混合物的 SLD。 - 对于 NSE 测量平均膜厚度波动(没有横向对比),请使用成分脂质的尾部除臭变体,即 DMPC-d54 (C36H18NO8PD54, 732.3 g/mol) 和 DSPC-d70 (C44H18NO8PD70, 860.1 g/mol)35,38, 这样mDMPC -d54 × 133.0 毫克和mDSPC-d70 × 67.0 毫克。此对比方案(图 2A,右面板)通过对比尾组 (NSLD = 6.4 × 10-6 +-2)与去质缓冲器(NSLD = 4.5 × 10-6 +-2)的散射信号进行对比匹配,从而能够检测膜厚度的波动。
- 对于某些膜隔间的厚度波动研究,例如,富含 DMPC 的矩阵, 使用第 1.2 步中描述的策略,用尾部解释的模拟(即 DMPC-d54)替换增殖的 DMPC 脂质,使 DSPC 丰富的域与去质缓冲区形成对比,并且主要散射信号来自尾部已脱释的 DMPC 富含矩阵的头部区域。
2. 为挤出准备脂质悬架
- 根据样本成分计算样本中每个成分的质量。根据经验,对于具有多种分子成分的样品,成分的质量由其摩尔质量Mwi给出,由其摩尔分数 Xi加权,并在所有成分上正常化,例如
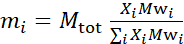 :Mtot 是总质量,设置在这里200毫克。请参阅上面的示例,用于具有不同解化方案的 DMPC-DSPC 脂质双层。
:Mtot 是总质量,设置在这里200毫克。请参阅上面的示例,用于具有不同解化方案的 DMPC-DSPC 脂质双层。 - 使用数字半微平衡,称量计算出的脂质质量(以及其他样品成分,如蛋白质、纳米粒子等),并将其添加到小瓶或圆底烧瓶中 , 记得事先称量小瓶或烧瓶。加入 1 mL 溶剂,通过手动在发动机罩内混合来溶解称重的组件。对于纯脂质样品,请使用氯仿或乙醇。对于具有额外非脂质组件(如纳米粒子)的样品,请选择分散所有组件的常见溶剂。
- 对于小脂质量(<10毫克),准备一个库存溶液和移液器所需的体积到混合物。
注意:不要添加过量的溶剂,因为这将显著减慢下面描述的溶剂干燥步骤。
- 对于小脂质量(<10毫克),准备一个库存溶液和移液器所需的体积到混合物。
- 将惰性气体(例如氮气、砷)轻轻流入小瓶中,同时以一个角度缓慢旋转小瓶,从而干燥发动机罩内的脂质溶液。将小瓶保持倾斜位置,在小瓶壁上形成干燥脂质的薄膜,甚至允许干燥。间歇性地将小瓶置于 35 °C 的水浴中,以规避蒸发介质冷却,从而减缓溶剂蒸发。
- 将小瓶在 +35 °C 的真空烤箱中过夜,以完全去除剩余溶剂。对于不饱和脂质,用惰性气体清除真空,以尽量减少氧化。
- 为确保完全去除溶剂,在脂质干燥后称量小瓶,并确认超出测量材料量的超量质量。这样做,在干燥后从测量的质量中减去小瓶的质量。如果质量超标,在真空下将样品再干燥 6 小时。根据需要重复此过程。
- 用 4 mL D2O 或 D2O 缓冲器将脂质薄膜水合,获得 50 毫克/mL 的脂质浓度。对于具有高过渡温度的脂质(如 DMPC-DSPC 混合物),将缓冲器加热到过渡温度(60 °C)以上,以确保均匀混合。
注:由于 NSE 实验需要相对较大的样本量 (+4 mL),因此考虑使用所需缓冲区的一半(即 2 mL)为样品保湿,以最大限度地减少每个样本的挤出次数(见第 3 节)。在这种情况下,添加缓冲后挤出的剩余一半。请注意,用于挤出的注射器容量限制在 1 mL 以内。因此,用4兆升缓冲器保湿需要四套挤出。 - 漩涡混合水合脂溶液,直到脂质薄膜完全溶解,不再可见小瓶的墙壁。在这个阶段,水合脂形成多拉梅拉囊泡和微米大小的多拉梅拉堆栈,悬浮物呈乳白色。
- 为了方便脂质堆栈的破裂和减少多重拉米亚,将水脂溶液小瓶放在实验室级冰柜(最好是 -80 °C 冰柜)中,直到完全冻结,然后将小瓶转移到 35 °C 的水浴中,直到脂质溶液完全解冻,从而执行五个冻结/解冻周期。漩涡解冻溶液,直到同质。再重复四次。
注意:或者,干冰浴可以通过将丙酮和干冰相结合来准备快速冷冻。
3. 水合脂溶液的挤出
- 使用两个膜支架之间的聚碳酸酯膜组装挤出器设置,并在两侧添加两个纸过滤器以提供额外的支持。使用孔径与目标脂膜大小相匹配的聚碳酸酯膜(NSE 实验的常见孔径为 50 nm 和 100 nm - 通常,直径为 100 nm 的脂质体允许较少约束膜波动,但较小的 50 nm 脂质体可用于曲率研究)。在完成装配和拧紧外部挤出器外壳之前,确保聚碳酸酯膜完全拉伸。
- 使用密封玻璃注射器通过膜组装几次,通过 +0.3 mL 的 D2O 或 D2O 缓冲,使聚碳酸酯膜水合。使用样品制备中使用的相同缓冲。离开它至少10分钟,然后完全吸出缓冲,然后再引入样品。
- 将 1 mL 气密注射器与准备好的脂质溶液一起填充,并插入挤出器的一端。然后,将空注射器插入另一端。一旦注射器连接到挤出器组件,将其放入挤出器块中。
- 如果挤出需要高温,如在过渡温度较高的饱和脂质(如 DSPC、Tm = 55 °C)的情况下),通过将加热块放在热板上或使用 图 3A中显示的循环浴池,预热脂质过渡温度(例如 60 °C)上方的挤出器加热块。
注:这一步骤对于确保脂质的均匀混合和避免在挤压过程中施加极端压力(可能会破裂聚碳酸酯膜)至关重要。对于过渡温度低(<25°C)的脂质样品,在室温下进行挤压。 - 要挤出脂质溶液,请将挤出器固定在可编程注射器泵上,并配有铝/钢框架, 如图 3A所示。对于温度控制的挤出,添加带流体通道的定制挤出器底座,并连接到循环水浴。
- 根据制造商的手册对注射器泵进行编程,以执行 15-20 个挤出周期。挤压时,脂质溶液的颜色从乳白色变为透明蛋白蓝色(图3B,C),表示最终的脂质大小小于可见光的波长,如预期的那样。对于图 3A中显示的注射器泵类型,请按照下面的步骤操作。
- 首先调整泵设置。按住 速率 按钮并输入挤出速率(50.99 mL/h),然后按 下直径 按钮并输入注射器直径(4.606 mm)。使用屏幕上每个数字下的向上箭头更改该数字值。
- 将带样品注射器的挤出器放在右边(见 图3A)。按 下取款 按钮,直到取款灯打开。 按"启动 "并等待样品分配到左侧(空)注射器。
- 在样品(右)注射器完全空之前,按下 停止 按钮。记录分配的体积并使用它来编程挤出周期。按住 速率 按钮,直到屏幕显示第 1 阶段 (PH:01)。按 下音量 按钮以输入之前记录的分配音量。在此阶段,确保取出灯关闭 - 这将样品分配在正确的方向。
- 再次按 下速率 按钮,使用最右向上箭头访问第 2 阶段(PH:02)。按 量 输入之前记录的分配卷的相同值。在此阶段,按下取款按钮,直到 取款 灯亮起 - 这将样品分配到左侧。
- 要重复此循环,请再次按 下速率 按钮,并使用最右向上箭头访问阶段 3 (PH:03)。按下 音量 按钮,直到 LP:SE 出现在屏幕上,并将其设置为 20。这是泵将执行的循环或重复数。最后,按 下速率 按钮,访问阶段 4 (PH:04),并按 下音量 按钮即可进入 停止 功能。泵现在设置为自动挤出。
- 按 开始 开始挤出周期。
- 将装有挤压脂质悬浮液的注射器清空,并准备储存或测量。对于熔化温度较高的脂质样品,将样品储存在流体相位过渡上方,直到测量。否则,请将样品保存在室温下。
- 不要冻结挤压的样品,因为冷冻会导致囊泡破裂(悬浮物将再次变成乳白色)。
4. NSE 对样本的测量和收集数据的减少
- 在 NSE 实验之前,使用可用方法从第 3.7 步开始描述挤出的脂质样本,以确保足够的样本质量。讨论部分包括可用于评估 NSE 实验的脂质悬浮质量的潜在焦化方法列表,例如大小分布、多重拉米、横向膜结构。
- 确定实验所需的 Q 范围和相应的仪器设置。对于脂质双层的弯曲刚度测量,请使用 +0.04 - 0.2) +-1 的Q 范围。对于膜厚度波动的研究,使用与膜厚度35、66、67对应的 +(0.04 -0.2)+ -1 的Q 范围。
注:在实验开始前与仪器科学家讨论实验设置。如前所述,样本的 SANS 定性是必要的,特别是如果无法获得散射信号的先前信息,如选择性脱水膜。或者,在 NSE 仪器上的有限 Q 范围上进行静态(也称为衍射)测量,并警告此类测量比 SANS 需要更长的时间。 - 使用注射器或转移移液器,将挤出的脂质悬浮液加载到 NSE 光束线可用的指定样本单元中。请注意,标准 NSE 样本细胞的厚度为 1、2、3 和 4 mm。选择细胞厚度,以优化散射信号,同时将不连贯的背景信号保持在合理的强度。
注:根据经验,使用1或2毫米路径长的样本细胞在去质缓冲区中产生脂质体 - 较厚的细胞可能导致难以纠正的多重散射效应。对于具有较高脱脂水平的脂质体(例如,尾部对比匹配的脂质体或带有单个增压传单的非对称脂质体),考虑使用较厚的样本细胞(例如,3 或 4 mm 路径长)来增强计数统计数据(如果样本数量较大,有时成本可能高得令人望而却步) - 为缓冲器准备相同的样本细胞。使用与脂质悬架相同的缓冲。对于强度正常化和背景 (BKG) 校正,缓冲器上的测量是必要的。
- 将样本细胞放在 NSE 光谱仪的样本支架中,对测量运行进行编程并收集回波数据。咨询仪器科学家有关首次使用 NSE 用户的测量编程。
- 执行数据减少所需的另外两组测量:分辨率(R)和传输(T)测量。
- 对弹性散射参考(例如碳)进行分辨率(R)测量 -在同一设置下运行;即与样品和缓冲器测量相同的波浪体和四倍。
- 对样品和缓冲器进行传输(T) 测量,以计算传输中子束的强度(参见下文第 4.9 步)。传输的计算方式是从样品或缓冲区分出的中子计数与开梁(即空样品位置)的中子计数之比。
- 使用执行测量的 NSE 光谱仪的专用数据减少软件来减少收集的数据。
注:不同的光谱仪可能会使用不同的软件或用户界面。以下是使用数据分析和可视化环境 (DAVE) 减少 NSE 数据的示例68 专门为 NIST 中子研究中心的 NSE 光谱仪编写的软件。- 打开 DAVE 软件,从数据减少菜单中选择 "减少 NSE 数据 "。将出现几个弹出窗口。
- 使用文件菜单上的"打开.echo 文件"将数据文件上传到不同的 Q 值上。这些文件与带有自旋回声信号的原始数据文件相对应,并在文件名中具有扩展。echo。文件上传完成后,文件将在可用数据集下显示。
- 右键单击所选文件,并根据其对应的测量结果将其标记:即,样品、细胞(用于空单元或缓冲器)或分辨率。
- 将探测器的尖刺分组在 2 x 2 中,以便使用 数据集 选项卡提高信号与噪声比。将相同的装箱应用到所有文件中;即分辨率、单元格和示例。
- 通过按键盘上的m键,检查所有像素组的数据,并屏蔽信号较差的组(见图 4B)。按"输入"以访问弹出窗口,以便对所有 Fourier 时间或随后的 Fourier 时间应用相同的面罩。在数据减少期间,这也可以在任何时候应用于单个像素。蒙面像素将变绿。
- 确保收集的数据以回声信号的形式出现,即在每个探测器像素上按相电流(见 图 4A)的 cosine 功能。
注:相流与中子自旋的入射角成正比:因此,通常将相流表示为 图 4A中显示的相位角度。对于脉冲源的测量,将额外的飞行计算时间应用于数据,以获取回声信号,作为中子脉冲中事件中子波长的函数。 - 首先安装分辨率文件。从上传的文件列表中选择一个分辨率文件,然后右键单击文件。选择适合操作:从弹出式菜单中选择适合回声(分辨率)。
- 确保回声信号的拟合产生一些拟合参数,包括第 4.8 步所需的参数 A。适合会使用以下表示方式自动执行。
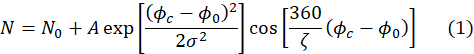
在这里,ζ回声信号的周期(即图4A中的宇宙函数),σ是由事件中子束的平均波长和波长扩散决定的高斯包络的宽度,+c是相流,+0是回波点,取决于中子50经历的场路径。有关样本的物理信息编码在方程中 cosine 函数的振幅A(1)中。
注:高斯信封的宽度基于仪器科学家预先确定的值,不应更改。其他参数是安装在每个像素上特定回波信号上的变量。 - 通过单击每个像素来检查拟合结果,以显示由此产生的拟合参数、拟合质量以及拟合的平均方形偏差。要检查整个探测器上每个拟合参数相关的错误,请选择 图像选项 ,然后选择感兴趣的拟合参数。这将生成一张地图,每个像素上都带有拟合参数的值。右键单击探测器图像。弹出窗口将显示所选拟合参数的差错栏图。
- 如果对特定像素的拟合不满意(例如,适合具有大误差条的参数会议),则将信号重新拟合到该特定像素上。选择该像素,按"拟合"选项卡,然后按"适合像素"。在安装选项卡中输入相(+0)和期间 (ζ) 的新起始参数,以获得更令人满意的适合。
注:将安装阶段绘制为傅立元时间的函数是有用的。为此,请前往主情节窗口,并选择 适合阶段诉傅立元时间。此绘图应平稳且连续。检查此图中的不连续性并重新安装其对应的像素。
- 通过从上传和标记的文件列表中选择相应的文件来减少样本或单元格文件。
- 检查所有像素,并掩盖那些与不良统计在步骤4.7.5中描述的。
- 右键单击文件并选择 适合操作:导入阶段(示例、单元格)。这将从分辨率文件中导入相位和应用面膜。
- 使用之前为分辨率文件描述的相同程序(步骤 4.7.8-4.7.10)安装回声信号。在安装示例和单元格文件时,不要更改从分辨率适合导入的周期值和回波相点。这些参数取决于工具设置,不应随示例而变化。
- 在开始数据缩减之前,输入所有数据文件的光束中心。选择数据文件,进入 一般 选项卡并输入 X 和 Y 光束中心值。这些值在实验期间记录。
- 一旦与示例、单元格和分辨率文件相适应,计算以后用于数据分析和解释的规范化中间散射函数。为此,请直接单击要从已安装文件列表中减少的示例文件,并从弹出式菜单中选择 "计算 I(Q)"。 窗口将显示分辨率和单元格(即缓冲)文件的条目选项以及 Q 弧数(参见步骤 4.9)。输入所有所需的信息后,按 下 OK 按钮。结果将出现在一个新的窗口中。
注:数据减少按以下方程进行,以获得规范化的中间散射函数69。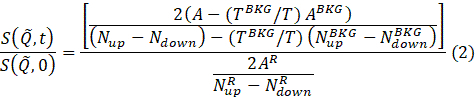
t是 Fourier 时间,N向上和向下 是非自旋翻转和自旋翻转配置中的中子计数(分别用π/2 翻转器和π翻转器关闭和打开)中的中子计数,以及超级脚本BKG和R,分别对应于第 4.4 步和 4.6 步中定义的背景和分辨率测量。请注意,光束极化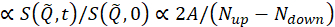 ,因此由于中子和样品之间的能量交换而改变自旋状态,检测到极化(来自统一)的下降。
,因此由于中子和样品之间的能量交换而改变自旋状态,检测到极化(来自统一)的下降。
- 最后,将探测器像素分组成图4B中显示的 Q弧,以获得标准化中间散射函数S(Q,t)/ S(Q,0) 的 Q 依赖性。这在技术上称为数据装箱,应明智地进行,即考虑到样本的计数统计数据和数据在分组像素上的预期标准偏差。
- 对于强散射样品,将探测器分成更多的Q弧,同时保持由此产生的中间散射函数S(Q,t)/S(Q,0)的合理误差条。这会产生更多的 Q 数据点,并且对于下面描述的数据分析过程非常重要。请注意,对于散射较弱的样品,过度装箱会导致衰变信号差,即S(Q,t)/ S(Q,0) 上的大误差条可能导致较大的不确定性。
5. 数据分析和解释
- 适合从上述数据减少中获得的规范化中间散射函数S(Q, t) / S(Q,0), 以 2/370的拉伸指数函数获得。
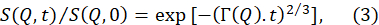
注:图5B提供了这些拟合的示例。S(Q,t) / S(Q, 0) 到方程 (3) 的适合产生 Q 依赖放松率Γ(Q)。 - 绘图Γ(Q)作为Q的函数,适合提取相关膜参数的合适模型。
结果
NSE 研究访问弯曲波动通常执行在 Q 范围 = (0.04 - 0.2) +-1。此 Q 范围对应膜厚度和脂质半径之间的中间长度刻度,弯曲动力学占主导地位。在扩展的 Q 范围上进行测量可以访问其他动态模式,包括脂质扩散和内膜动力学。有关 NSE 访问的膜动力学交叉的更多详细信息,请查看这些相关出版物25,71。重要的是要强调,NSE信号是成正比
讨论
NSE 是一种功能强大且独特的技术,可测量不同条件下脂质膜的中等动力学。NSE 的有效利用取决于样品质量、中子对比度以及可用于特定样本的可访问动力学范围。因此,要成功进行 NSE 实验和收集高质量数据,需要几个关键步骤。确保在 NSE 实验中有效利用中子束时间的一个关键步骤是在 NSE 实验之前使用基于实验室的方法对脂质悬浮进行特征描述。对于外显子,挤出脂质体的大小分布(或扩散?...
披露声明
作者声明没有利益冲突,也没有什么可透露的。
致谢
阿什卡尔感谢长高,洛杉矶斯廷加丘和P.佐尔涅尔丘克进行了许多有益的讨论,并经常协助NSE在各自的光束线上进行实验。作者承认在NIST和ORNL使用中子自旋回波光谱仪。NIST 的 NSE 光谱仪由高分辨率中子散射中心提供支持,该中心是国家标准与技术研究所和国家科学基金会根据第 11 号协议建立的伙伴关系。DMR-1508249。ORNL 喷射中子源的 NSE 光谱仪由美国能源部基础能源科学办公室科学用户设施司提供支持。橡树岭国家实验室由美国国防部合同第一号公司 UT-Battelle 管理。DE-AC05-00OR22725。
材料
| Name | Company | Catalog Number | Comments |
| Chloroform (biotech grade) | Sigma Aldrich | 496189 | Biotech. grade, ≥99.8%, contains 0.5-1.0% ethanol as stabilizer |
| Circulating water bath | Julabo | SE-12 | Heating Circulator with smart pump, programmable temperature settings, and external sensor connection for measurement and control |
| Deuterium Oxide | Cambridge Isotopes Laboratories | DLM-4 | Deuterated water; Heavy water (D2O) (D, 99.9%) |
| Digital Semi-Microbalance | Mettler Toledo | MS105 | Semi-micro balance with 120 g capacity, 0.01 mg readability, high resolution weighing cell, ergonomic doors, and pipette-check application |
| Ethanol (molecular biology grade) | Sigma Aldrich | E7023 | 200 proof ethanol for molecular biology applications |
| Glass Pipets | VWR | 36360-536 | Disposable Soda Lime glass Pasteur pipets |
| Glass Vials | Thermo Scientific | B7990-1 | Borosilicate glass vials with PTFE/Silione septum caps |
| Lab grade freezer | Fisher Scientific | IU2886D | Ultra-low temprature freezer (-86 to -50 C) for long-term storage of lipids and proteins |
| Lipids (protaited or perdeuterated) | Avanti Polar Lipids | varies by lipid | Lipids can be purchased from Avanti in powder form or in a chloroform solution with the required amounts and deuteration schemes. |
| Millipore water purifier | Millipore Sigma | ZRQSVP3US | Direct-Q® 3 UV Water Purification System which deliver both pure and ultrapure water with a built-in UV lamp to reduce the levels of organics for biological applications |
| Mini Extruder Set | Avanti Polar Lipids | 610020 | Mini-extruder set includes mini-extruder, heating block, 2 GasTight Syringes, and 2 O-rings, Polycarbonate Membranes, and Filter Supports |
| Quick Connect Fittings | Grainger | 2YDA1 and 2YDA7 | Push-button tube fittings for QuickConnect water circulation applications, e.g. high temperature vesicle extrusion |
| Syringe Pump | SyringePump.com | New Era-1000 | Fully programmable syringe pump for infusion and withdrawal; programs up to 41 pumping phases with adjustable pumping rates, dispensed volumes, and extrusion cycles |
| Ultrasonic bath | Fisher Scientific | CPX2800 | Temperature controlled ultra sonic bath with programmable functionality for degassing and ultrasonic applications |
| Vacuum Oven | Thermo Scientific | 3608 | 0.7 cu ft vaccum oven with built-in-high-limit thermostat guards against overheating |
| Vortex Mixer | Fisher Scientific | 02-215-414 | Variable speed, analog control that allows low rpm start-up for gentle shaking or high-speed mixing for vigorous vortexing of samples |
参考文献
- Singer, S. J., Nicolson, G. L. The fluid mosaic model of the structure of cell membranes. Science. 175 (4023), 720-731 (1972).
- Andersen, O. S., Koeppe, R. E. Bilayer thickness and membrane protein function: an energetic perspective. Annual Review of Biophysics and Biomolecular Structure. 36, 107-130 (2007).
- Lundbæk, J. A., Collingwood, S. A., Ingólfsson, H. I., Kapoor, R., Andersen, O. S. Lipid bilayer regulation of membrane protein function: gramicidin channels as molecular force probes. Journal of The Royal Society Interface. 7 (44), 373-395 (2010).
- Bradley, R. P., Radhakrishnan, R. Curvature-undulation coupling as a basis for curvature sensing and generation in bilayer membranes. Proceedings of the National Academy of Sciences of the United States of America. 113 (35), 117-124 (2016).
- Perozo, E., Cortes, D. M., Sompornpisut, P., Kloda, A., Martinac, B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 418 (6901), 942-948 (2002).
- Jensen, M. &. #. 2. 1. 6. ;., Mouritsen, O. G. Lipids do influence protein function-the hydrophobic matching hypothesis revisited. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1666 (1-2), 205-226 (2004).
- Rajendran, L., Simons, K. Lipid rafts and membrane dynamics. Journal of Cell Science. 118 (6), 1099-1102 (2005).
- Katchalsky, A., Spangler, R. Dynamics of membrane processes. Quarterly Reviews of Biophysics. 1 (2), 127-175 (1968).
- Rheinstädter, M. C. Collective molecular dynamics in proteins and membranes (Review). Biointerphases. 3 (2), 83-90 (2008).
- Fujiwara, T., Ritchie, K., Murakoshi, H., Jacobson, K., Kusumi, A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. The Journal of Cell Biology. 157 (6), 1071-1082 (2002).
- Hac, A. E., Seeger, H. M., Fidorra, M., Heimburg, T. Diffusion in two-component lipid membranes--a fluorescence correlation spectroscopy and monte carlo simulation study. Biophysical Journal. 88 (1), 317-333 (2005).
- Heinrich, M., Tian, A., Esposito, C., Baumgart, T. Dynamic sorting of lipids and proteins in membrane tubes with a moving phase boundary. Proceedings of the National Academy of Sciences of the United States of America. 107 (16), 7208-7213 (2010).
- Hormel, T. T., Kurihara, S. Q., Brennan, M. K., Wozniak, M. C., Parthasarathy, R. Measuring lipid membrane viscosity using rotational and translational probe diffusion. Physical Review Letters. 112 (18), 188101 (2014).
- Dimova, R. Recent developments in the field of bending rigidity measurements on membranes. Advances in Colloid and Interface Science. 208, 225-234 (2014).
- Bassereau, P., Sorre, B., Lévy, A. Bending lipid membranes: Experiments after W. Helfrich's model. Advances in Colloid and Interface Science. 208, 47-57 (2014).
- Monzel, C., Sengupta, K. Measuring shape fluctuations in biological membranes. Journal of Physics D: Applied Physics. 49 (24), 243002 (2016).
- Deserno, M. Mesoscopic membrane physics: concepts, simulations, and selected applications. Macromolecular Rapid Communications. 30 (9-10), 752-771 (2009).
- Reynwar, B. J., et al. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 447 (7143), 461-464 (2007).
- Haswell, E. S., Phillips, R., Rees, D. C. Mechanosensitive channels: what can they do and how do they do it. Structure. 19 (10), 1356-1369 (2011).
- Phillips, R., Ursell, T., Wiggins, P., Sens, P. Emerging roles for lipids in shaping membrane-protein function. Nature. 459 (7245), 379-385 (2009).
- Dill, K. A., Chan, H. S. From Levinthal to pathways to funnels. Nature Structural Biology. 4 (1), 10-19 (1997).
- Henzler-Wildman, K., Kern, D. Dynamic personalities of proteins. Nature. 450 (7172), 964-972 (2007).
- Grimaldo, M., Roosen-Runge, F., Zhang, F., Schreiber, F., Seydel, T. Dynamics of proteins in solution. Quarterly Reviews of Biophysics. 52, 7 (2019).
- Lyman, E., Hsieh, C. -. L., Eggeling, C. From dynamics to membrane organization: experimental breakthroughs occasion a "modeling manifesto". Biophysical Journal. 115 (4), 595-604 (2018).
- Arriaga, L. R., et al. Dissipative curvature fluctuations in bilayer vesicles: Coexistence of pure-bending and hybrid curvature-compression modes. The European Physical Journal. E, Soft Matter. 31 (1), 105-113 (2010).
- Honerkamp-Smith, A. R., Veatch, S. L., Keller, S. L. An introduction to critical points for biophysicists; observations of compositional heterogeneity in lipid membranes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1788 (1), 53-63 (2009).
- Veatch, S. L., Keller, S. L. Organization in lipid membranes containing cholesterol. Physical Review Letters. 89 (26), 268101 (2002).
- Heberle, F. A., et al. Bilayer thickness mismatch controls domain size in model membranes. Journal of the American Chemical Society. 135 (18), 6853-6859 (2013).
- Nickels, J. D., et al. The in vivo structure of biological membranes and evidence for lipid domains. PLOS Biology. 15 (5), 2002214 (2017).
- Simons, K., Ikonen, E. Functional rafts in cell membranes. Nature. 387 (6633), 569-572 (1997).
- van Meer, G., Voelker, D. R., Feigenson, G. W. Membrane lipids: where they are and how they behave. Nature Reviews. Molecular Cell Biology. 9 (2), 112-124 (2008).
- Liu, S. -. L., et al. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nature Chemical Biology. 13, 268 (2016).
- Rothman, J., Lenard, J. Membrane asymmetry. Science. 195 (4280), 743-753 (1977).
- Ashkar, R., et al. Neutron scattering in the biological sciences: progress and prospects. Acta Crystallographica Section D. 74 (12), 1129-1168 (2018).
- Woodka, A. C., Butler, P. D., Porcar, L., Farago, B., Nagao, M. Lipid bilayers and membrane dynamics: insight into thickness fluctuations. Physical Review Letters. 109 (5), 058102 (2012).
- Chakraborty, S., et al. How cholesterol stiffens unsaturated lipid membranes. Proceedings of the National Academy of Sciences of the United States of America. 117 (36), 21896-21905 (2020).
- Arriaga, L. R., et al. Stiffening effect of cholesterol on disordered lipid phases: a combined neutron spin echo + dynamic light scattering analysis of the bending elasticity of large unilamellar vesicles. Biophysical Journal. 96 (9), 3629-3637 (2009).
- Nagao, M., Kelley, E. G., Ashkar, R., Bradbury, R., Butler, P. D. Probing elastic and viscous properties of phospholipid bilayers using neutron spin echo spectroscopy. The Journal of Physical Chemistry Letters. 8 (19), 4679-4684 (2017).
- Kelley, E. G., Butler, P. D., Ashkar, R., Bradbury, R., Nagao, M. Scaling relationships for the elastic moduli and viscosity of mixed lipid membranes. Proceedings of the National Academy of Sciences of the United States of America. 117 (38), 23365-23373 (2020).
- Rickeard, B. W., et al. Transverse lipid organization dictates bending fluctuations in model plasma membranes. Nanoscale. 12 (3), 1438-1447 (2020).
- Nickels, J. D., et al. Mechanical properties of nanoscopic lipid domains. Journal of the American Chemical Society. 137 (50), 15772-15780 (2015).
- Mezei, F. Neutron spin echo: A new concept in polarized thermal neutron techniques. Zeitschrift für Physik A Hadrons and Nuclei. 255 (2), 146-160 (1972).
- Hayter, J. B., Penfold, J. Neutron spin-echo integral transform spectroscopy. Zeitschrift für Physik B Condensed Matter. 35 (2), 199-205 (1979).
- Monkenbusch, M., Richter, D., Imae, T., Kanaya, T., Furusaka, M., Torikai, N. . Neutrons in Soft Matter. , 147-182 (2011).
- Pynn, R., Mezei, F., Pappas, C., Gutberlet, T. . Neutron Spin Echo. , 159-177 (2003).
- Holderer, O., et al. The JCNS neutron spin-echo spectrometer J-NSE at the FRM II. Measurement Science and Technology. 19 (3), 034022 (2008).
- Schleger, P., et al. The long-wavelength neutron spin-echo spectrometer IN15 at the Institut Laue-Langevin. Physica B: Condensed Matter. 241-243, 164-165 (1997).
- Holderer, O., Zolnierczuk, P., Pasini, S., Stingaciu, L., Monkenbusch, M. A better view through new glasses: Developments at the Jülich neutron spin echo spectrometers. Physica B: Condensed Matter. 562, 9-12 (2019).
- Farago, B., et al. The IN15 upgrade. Neutron News. 26 (3), 15-17 (2015).
- Ashkar, R. Selective dynamics in polymeric materials: Insights from quasi-elastic neutron scattering spectroscopy. Journal of Applied Physics. 127 (15), 151101 (2020).
- Pasini, S., Holderer, O., Kozielewski, T., Richter, D., Phoenix Monkenbusch, M. J-NSE- Phoenix, a neutron spin-echo spectrometer with optimized superconducting precession coils at the MLZ in Garching. Review of Scientific Instruments. 90 (4), 043107 (2019).
- Svergun, D. I., Koch, M. H. J., Timmins, P. A., May, R. P. . Small Angle X-Ray and Neutron Scattering from Solutions of Biological Macromolecules. , (2013).
- Eicher, B., et al. Joint small-angle X-ray and neutron scattering data analysis of asymmetric lipid vesicles. Journal of Applied Crystallography. 50 (2), 419-429 (2017).
- Heberle, F. A., et al. Model-based approaches for the determination of lipid bilayer structure from small-angle neutron and X-ray scattering data. European Biophysics Journal. 41 (10), 875-890 (2012).
- Jaksch, S., Koutsioubas, A., Mattauch, S., Holderer, O., Frielinghaus, H. Long-range excitations in phospholipid membranes. Chemistry and Physics of Lipids. 225, 104788 (2019).
- Jaksch, S., et al. Influence of ibuprofen on phospholipid membranes. Physical Review E. 91 (2), 022716 (2015).
- Armstrong, C. L., et al. Effect of cholesterol on the lateral nanoscale dynamics of fluid membranes. European Biophysics Journal. 41 (10), 901-913 (2012).
- Rheinstädter, M. C., Häußler, W., Salditt, T. Dispersion relation of lipid membrane shape fluctuations by neutron spin-echo spectrometry. Physical Review Letters. 97 (4), 048103 (2006).
- Armstrong, C. L., Häußler, W., Seydel, T., Katsaras, J., Rheinstädter, M. C. Nanosecond lipid dynamics in membranes containing cholesterol. Soft Matter. 10 (15), 2600-2611 (2014).
- Nickels, J. D., et al. Lipid rafts: buffers of cell membrane physical properties. The Journal of Physical Chemistry B. 123 (9), 2050-2056 (2019).
- Michonova-Alexova, E. I., Sugár, I. P. Component and state separation in DMPC/DSPC lipid bilayers: a Monte Carlo simulation study. Biophysical Journal. 83 (4), 1820-1833 (2002).
- Sugár, I. P., Thompson, T. E., Biltonen, R. L. Monte Carlo simulation of two-component bilayers: DMPC/DSPC mixtures. Biophysical Journal. 76 (4), 2099-2110 (1999).
- Mabrey, S., Sturtevant, J. M. Investigation of phase transitions of lipids and lipid mixtures by sensitivity differential scanning calorimetry. Proceedings of the National Academy of Sciences. 73 (11), 3862-3866 (1976).
- . Neutron activation and scattering calculator Available from: https://www.ncnr.nist.gov/resources/activation/ (2021)
- Scott, H. L., et al. On the mechanism of bilayer separation by extrusion, or why your LUVs are not really unilamellar. Biophysical Journal. 117 (8), 1381-1386 (2019).
- Ashkar, R., et al. Tuning membrane thickness fluctuations in model lipid bilayers. Biophysical Journal. 109 (1), 106-112 (2015).
- Carrillo, J. -. M. Y., Katsaras, J., Sumpter, B. G., Ashkar, R. A computational approach for modeling neutron scattering data from lipid bilayers. Journal of Chemical Theory and Computation. 13 (2), 916-925 (2017).
- Azuah, R. T. DAVE: a comprehensive software suite for the reduction, visualization, and analysis of low energy neutron spectroscopic data. Journal of Research of the National Institute of Standards and Technology. 114 (6), 341-358 (2009).
- Van Hove, L. Correlations in space and time and born approximation scattering in systems of interacting particles. Physical Review. 95 (1), 249-262 (1954).
- Zilman, A. G., Granek, R. Undulations and dynamic structure factor of membranes. Physical Review Letters. 77 (23), 4788-4791 (1996).
- Kelley, E. G., Butler, P. D., Nagao, M. . Collective dynamics in model biological membranes measured by neutron spin echo spectroscopy. , 131-176 (2019).
- Zheng, Y., Michihiro, N., Dobrin, P. B. Bending elasticity of saturated and monounsaturated phospholipid membranes studied by the neutron spin echo technique. Journal of Physics: Condensed Matter. 21 (15), 155104 (2009).
- Sharma, V. K., Qian, S. Effect of an antimicrobial peptide on lateral segregation of lipids: a structure and dynamics study by neutron scattering. Langmuir. 35 (11), 4152-4160 (2019).
- Boggara, M. B., Faraone, A., Krishnamoorti, R. Effect of pH and Ibuprofen on the Phospholipid Bilayer Bending Modulus. The Journal of Physical Chemistry B. 114 (24), 8061-8066 (2010).
- Lee, J. -. H., et al. Thermal fluctuation and elasticity of lipid vesicles interacting with pore-forming peptides. Physical Review Letters. 105 (3), 038101 (2010).
- Chakraborty, S., Abbasi, A., Bothun, G. D., Nagao, M., Kitchens, C. L. Phospholipid bilayer softening due to hydrophobic gold nanoparticle inclusions. Langmuir. 34 (44), 13416-13425 (2018).
- Hoffmann, I., et al. Softening of phospholipid membranes by the adhesion of silica nanoparticles - as seen by neutron spin-echo (NSE). Nanoscale. 6 (12), 6945-6952 (2014).
- Watson, M. C., Brown, F. L. H. Interpreting membrane scattering experiments at the mesoscale: the contribution of dissipation within the bilayer. Biophysical Journal. 98 (6), 9-11 (2010).
- Seifert, U., Langer, S. A. Viscous modes of fluid bilayer membranes. Europhysics Letters (EPL). 23 (1), 71-76 (1993).
- Bingham, R. J., Smye, S. W., Olmsted, P. D. Dynamics of an asymmetric bilayer lipid membrane in a viscous solvent. EPL (Europhysics Letters). 111 (1), 18004 (2015).
- Rawicz, W., Olbrich, K. C., McIntosh, T., Needham, D., Evans, E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophysical Journal. 79 (1), 328-339 (2000).
- Doktorova, M., LeVine, M. V., Khelashvili, G., Weinstein, H. A new computational method for membrane compressibility: bilayer mechanical thickness revisited. Biophysical Journal. 116 (3), 487-502 (2019).
- Evans, E., Needham, D. Physical properties of surfactant bilayer membranes: thermal transitions, elasticity, rigidity, cohesion and colloidal interactions. The Journal of Physical Chemistry. 91 (16), 4219-4228 (1987).
- Lesieur, S., Grabielle-Madelmont, C., Paternostre, M. T., Ollivon, M. Size analysis and stability study of lipid vesicles by high-performance gel exclusion chromatography, turbidity, and dynamic light scattering. Analytical Biochemistry. 192 (2), 334-343 (1991).
- Heberle, F. A., et al. Direct label-free imaging of nanodomains in biomimetic and biological membranes by cryogenic electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 117 (33), 19943-19952 (2020).
- Cornell, C. E., Mileant, A., Thakkar, N., Lee, K. K., Keller, S. L. Direct imaging of liquid domains in membranes by cryo-electron tomography. Proceedings of the National Academy of Sciences of the United States of America. 117 (33), 19713-19719 (2020).
- Yao, X., Fan, X., Yan, N. Cryo-EM analysis of a membrane protein embedded in the liposome. Proceedings of the National Academy of Sciences of the United States of America. 117 (31), 18497-18503 (2020).
- Kučerka, N., Nieh, M. -. P., Katsaras, J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1808 (11), 2761-2771 (2011).
- Nielsen, J. E., Bjørnestad, V. A., Lund, R. Resolving the structural interactions between antimicrobial peptides and lipid membranes using small-angle scattering methods: the case of indolicidin. Soft Matter. 14 (43), 8750-8763 (2018).
- Kučerka, N., et al. Lipid bilayer structure determined by the simultaneous analysis of neutron and X-ray scattering data. Biophysical Journal. 95 (5), 2356-2367 (2008).
- Kelley, E. G., Butler, P. D., Nagao, M. Scaling of lipid membrane rigidity with domain area fraction. Soft Matter. 15 (13), 2762-2767 (2019).
- Brüning, B. -. A., et al. Bilayer undulation dynamics in unilamellar phospholipid vesicles: Effect of temperature, cholesterol and trehalose. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1838 (10), 2412-2419 (2014).
- Kučerka, N., et al. Areas of monounsaturated diacylphosphatidylcholines. Biophysical Journal. 97 (7), 1926-1932 (2009).
- Sharma, V. K., Mamontov, E., Anunciado, D. B., O'Neill, H., Urban, V. S. Effect of antimicrobial peptide on the dynamics of phosphocholine membrane: role of cholesterol and physical state of bilayer. Soft Matter. 11 (34), 6755-6767 (2015).
- Kelley, E. G., Butler, P. D., Nagao, M. Collective dynamics in lipid membranes containing transmembrane peptides. Soft Matter. , (2021).
- Yu, J., et al. Structure and dynamics of lipid membranes interacting with antivirulence end-phosphorylated polyethylene glycol block copolymers. Soft Matter. 16 (4), 983-989 (2020).
- Stingaciu, L. -. R., et al. Revealing the dynamics of thylakoid membranes in living cyanobacterial cells. Scientific Reports. 6 (1), 19627 (2016).
- Stingaciu, L. -. R., O'Neill, H. M., Liberton, M., Pakrasi, H. B., Urban, V. S. Influence of chemically disrupted photosynthesis on cyanobacterial thylakoid dynamics in synechocystis sp. PCC 6803. Scientific Reports. 9 (1), 5711 (2019).
- Miller, I. R. Energetics of fluctuation in lipid bilayer thickness. Biophysical Journal. 45 (3), 643-644 (1984).
- Nagao, M. Observation of local thickness fluctuations in surfactant membranes using neutron spin echo. Physical Review E. 80 (3), 031606 (2009).
Erratum
Formal Correction: Erratum: Neutron Spin Echo Spectroscopy as a Unique Probe for Lipid Membrane Dynamics and Membrane-Protein Interactions
Posted by JoVE Editors on 8/06/2021. Citeable Link.
An erratum was issued for: Neutron Spin Echo Spectroscopy as a Unique Probe for Lipid Membrane Dynamics and Membrane-Protein Interactions. The Introduction, Protocol, and Representative Results sections have been updated.
In the Introduction, the fith pargraph was updated from:
Besides direct access to the length and time scale of membrane dynamics, NSE has the inherent capabilities of neutron isotope sensitivity52. Specifically, the ability of neutrons to interact differently with the isotopes of hydrogen, the most abundant element in biological systems, results in a different neutron scattering length density,34 or NSLD (the equivalent of the optical index of refraction50), when protium is substituted by deuterium. This enables an approach known as contrast variation, which is commonly used to highlight specific membrane features or conceal others — the latter scenario is referred to as contrast matching. A frequent application of contrast variation/matching is the substitution of water (NSLD = -0.56 × 10-6 Å-2) by heavy water or D2O (NSLD = 6.4 × 10-6 Å-2) to amplify the neutron signal from protiated lipid membranes (NSLD ~ 2 × 10-6 Å-2). This approach is highly effective in studies of membrane structure because the penetration of D2O into the headgroup region of the membrane allows accurate determination of the membrane thicknesses (see Figure 2A, left panel) and of the location of different lipid subgroups when more sophisticated models are applied53,54. This paper highlights some examples on the use of contrast variation for studies of collective dynamics in biomimetic membranes and select membrane features.
to:
Besides direct access to the length and time scale of membrane dynamics, NSE has the inherent capabilities of neutron isotope sensitivity52. Specifically, the ability of neutrons to interact differently with the isotopes of hydrogen, the most abundant element in biological systems, results in a different neutron scattering length density,34 or NSLD (the equivalent of the optical index of refraction50), when protium is substituted by deuterium. This enables an approach known as contrast variation, which is commonly used to highlight specific membrane features or conceal others — the latter scenario is referred to as contrast matching. A frequent application of contrast variation/matching is the substitution of water (NSLD = -0.56 × 10-6 Å-2) by heavy water or D2O (NSLD = 6.4 × 10-6 Å-2) to amplify the neutron signal from protiated lipid membranes (NSLD ~ 0 × 10-6 Å-2). This approach is highly effective in studies of membrane structure because the penetration of D2O into the headgroup region of the membrane allows accurate determination of the membrane thicknesses (see Figure 2A, left panel) and of the location of different lipid subgroups when more sophisticated models are applied53,54. This paper highlights some examples on the use of contrast variation for studies of collective dynamics in biomimetic membranes and select membrane features.
In the Protocol, step 1.1 was updated from:
For bending fluctuation measurements, make fully protiated liposomes in D2O (D 99.9%) or D2O-buffer (e.g., phosphate buffer prepared with D2O instead of H2O). Use fully protiated DMPC (C36H72NO8P) and DSPC (C44H88NO8P) with 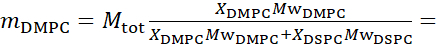 133.4 mg, where XDMPC and XDSPC are the mole fractions of DMPC and DSPC, here set to 0.7 and 0.3, respectively, and MwDMPC and MwDSPC are the molar weights given by 677.9 g/mol and 790.1 g/mol, respectively. Similarly, mDSPC = 66.6 mg. This deuteration scheme increases the scattering contrast between the membrane (NSLD ~ 2 × 10-6 Å-2) and the deuterated buffer (NSLD ~ 6.4 × 10-6 Å-2) and amplifies the signal from membrane undulations (see Figure 2A left panel).
133.4 mg, where XDMPC and XDSPC are the mole fractions of DMPC and DSPC, here set to 0.7 and 0.3, respectively, and MwDMPC and MwDSPC are the molar weights given by 677.9 g/mol and 790.1 g/mol, respectively. Similarly, mDSPC = 66.6 mg. This deuteration scheme increases the scattering contrast between the membrane (NSLD ~ 2 × 10-6 Å-2) and the deuterated buffer (NSLD ~ 6.4 × 10-6 Å-2) and amplifies the signal from membrane undulations (see Figure 2A left panel).
to:
For bending fluctuation measurements, make fully protiated liposomes in D2O (D 99.9%) or D2O-buffer (e.g., phosphate buffer prepared with D2O instead of H2O). Use fully protiated DMPC (C36H72NO8P) and DSPC (C44H88NO8P) with 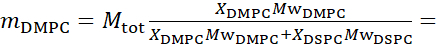 133.4 mg, where XDMPC and XDSPC are the mole fractions of DMPC and DSPC, here set to 0.7 and 0.3, respectively, and MwDMPC and MwDSPC are the molar weights given by 677.9 g/mol and 790.1 g/mol, respectively. Similarly, mDSPC = 66.6 mg. This deuteration scheme increases the scattering contrast between the membrane (NSLD ~ 0 × 10-6 Å-2) and the deuterated buffer (NSLD ~ 6.4 × 10-6 Å-2) and amplifies the signal from membrane undulations (see Figure 2A left panel).
133.4 mg, where XDMPC and XDSPC are the mole fractions of DMPC and DSPC, here set to 0.7 and 0.3, respectively, and MwDMPC and MwDSPC are the molar weights given by 677.9 g/mol and 790.1 g/mol, respectively. Similarly, mDSPC = 66.6 mg. This deuteration scheme increases the scattering contrast between the membrane (NSLD ~ 0 × 10-6 Å-2) and the deuterated buffer (NSLD ~ 6.4 × 10-6 Å-2) and amplifies the signal from membrane undulations (see Figure 2A left panel).
In the Representative Results, the fist pagargaph was updted from:
NSE studies accessing bending fluctuations are typically performed over a Q-range of ~ (0.04 - 0.2) Å-1. This Q-range corresponds to intermediate length scales between the membrane thickness and the liposomal radius, where bending dynamics dominate. Measurement over an extended Q-range can give access to additional dynamic modes, including liposomal diffusion and intramembrane dynamics. For more details on the cross-over in membrane dynamics accessed by NSE, check these relevant publications25,71. It is important to emphasize that NSE signals are proportional to: 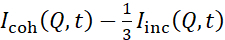 , where Icoh and Iinc are, respectively, the coherent and incoherent scattering intensity from the sample. Therefore, it is advisable to prepare NSE liposomal samples in deuterated buffers (i.e., buffers prepared with D2O instead of H2O) to minimize the incoherent scattering signal, mainly contributed by the hydrogen content of the sample. However, in some cases intermediate deuteration schemes (i.e., using mixtures of D2O and H2O) might be necessary to obtain optimal contrast conditions. Typically, NSE measurements of membrane bending fluctuations are performed on fully protiated liposomes in deuterated buffer, referred to as fully contrasted liposomes in Figure 5. This deuteration scheme results in a large NSLD difference between the membrane core (~2 × 10-6 Å-2) and its deuterated fluid environment (~6.4 × 10-6 Å-2), which significantly enhances the scattering signal from the liposomal membranes and improves the measurement statistics of bending dynamics. This contrast scheme (Figure 2A left panel) is frequently utilized in studies of bending rigidity of lipid membranes with single38,72 and multiple39,66 lipid components and in studies of membrane softening/stiffening by biological inclusions (e.g., cholesterol, drug molecules, peptides/proteins)36,37,73,74,75, and synthetic additives (e.g., nanoparticles)76,77.
, where Icoh and Iinc are, respectively, the coherent and incoherent scattering intensity from the sample. Therefore, it is advisable to prepare NSE liposomal samples in deuterated buffers (i.e., buffers prepared with D2O instead of H2O) to minimize the incoherent scattering signal, mainly contributed by the hydrogen content of the sample. However, in some cases intermediate deuteration schemes (i.e., using mixtures of D2O and H2O) might be necessary to obtain optimal contrast conditions. Typically, NSE measurements of membrane bending fluctuations are performed on fully protiated liposomes in deuterated buffer, referred to as fully contrasted liposomes in Figure 5. This deuteration scheme results in a large NSLD difference between the membrane core (~2 × 10-6 Å-2) and its deuterated fluid environment (~6.4 × 10-6 Å-2), which significantly enhances the scattering signal from the liposomal membranes and improves the measurement statistics of bending dynamics. This contrast scheme (Figure 2A left panel) is frequently utilized in studies of bending rigidity of lipid membranes with single38,72 and multiple39,66 lipid components and in studies of membrane softening/stiffening by biological inclusions (e.g., cholesterol, drug molecules, peptides/proteins)36,37,73,74,75, and synthetic additives (e.g., nanoparticles)76,77.
to:
NSE studies accessing bending fluctuations are typically performed over a Q-range of ~ (0.04 - 0.2) Å-1. This Q-range corresponds to intermediate length scales between the membrane thickness and the liposomal radius, where bending dynamics dominate. Measurement over an extended Q-range can give access to additional dynamic modes, including liposomal diffusion and intramembrane dynamics. For more details on the cross-over in membrane dynamics accessed by NSE, check these relevant publications25,71. It is important to emphasize that NSE signals are proportional to: 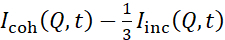 , where Icoh and Iinc are, respectively, the coherent and incoherent scattering intensity from the sample. Therefore, it is advisable to prepare NSE liposomal samples in deuterated buffers (i.e., buffers prepared with D2O instead of H2O) to minimize the incoherent scattering signal, mainly contributed by the hydrogen content of the sample. However, in some cases intermediate deuteration schemes (i.e., using mixtures of D2O and H2O) might be necessary to obtain optimal contrast conditions. Typically, NSE measurements of membrane bending fluctuations are performed on fully protiated liposomes in deuterated buffer, referred to as fully contrasted liposomes in Figure 5. This deuteration scheme results in a large NSLD difference between the membrane core (~0 × 10-6 Å-2) and its deuterated fluid environment (~6.4 × 10-6 Å-2), which significantly enhances the scattering signal from the liposomal membranes and improves the measurement statistics of bending dynamics. This contrast scheme (Figure 2A left panel) is frequently utilized in studies of bending rigidity of lipid membranes with single38,72 and multiple39,66 lipid components and in studies of membrane softening/stiffening by biological inclusions (e.g., cholesterol, drug molecules, peptides/proteins)36,37,73,74,75, and synthetic additives (e.g., nanoparticles)76,77.
, where Icoh and Iinc are, respectively, the coherent and incoherent scattering intensity from the sample. Therefore, it is advisable to prepare NSE liposomal samples in deuterated buffers (i.e., buffers prepared with D2O instead of H2O) to minimize the incoherent scattering signal, mainly contributed by the hydrogen content of the sample. However, in some cases intermediate deuteration schemes (i.e., using mixtures of D2O and H2O) might be necessary to obtain optimal contrast conditions. Typically, NSE measurements of membrane bending fluctuations are performed on fully protiated liposomes in deuterated buffer, referred to as fully contrasted liposomes in Figure 5. This deuteration scheme results in a large NSLD difference between the membrane core (~0 × 10-6 Å-2) and its deuterated fluid environment (~6.4 × 10-6 Å-2), which significantly enhances the scattering signal from the liposomal membranes and improves the measurement statistics of bending dynamics. This contrast scheme (Figure 2A left panel) is frequently utilized in studies of bending rigidity of lipid membranes with single38,72 and multiple39,66 lipid components and in studies of membrane softening/stiffening by biological inclusions (e.g., cholesterol, drug molecules, peptides/proteins)36,37,73,74,75, and synthetic additives (e.g., nanoparticles)76,77.
In the Representative Reults, Figure 2 was updated from:
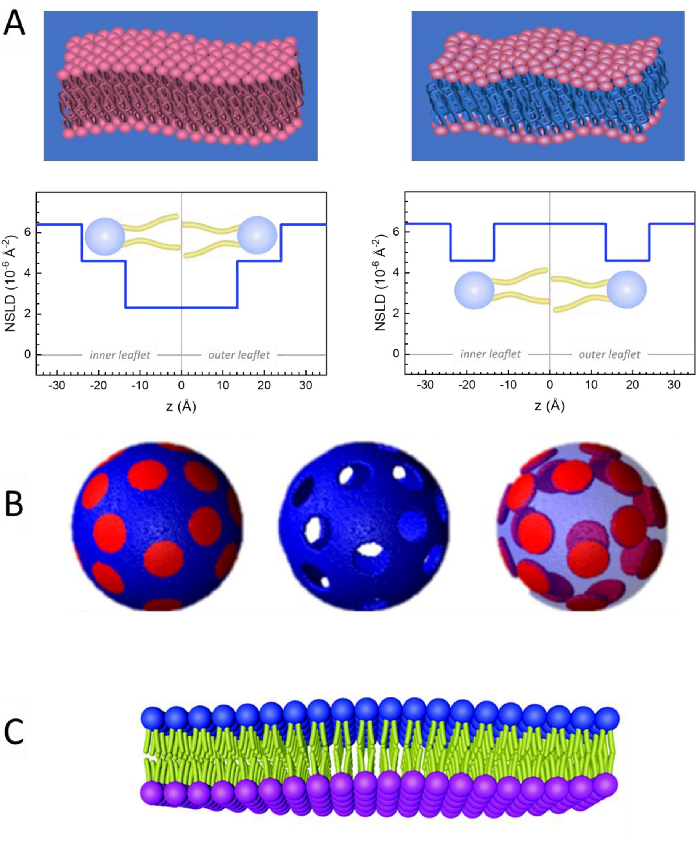
Figure 2: Examples of possible deuteration schemes in NSE experiments on lipid membranes. (A) Left: Fully contrasted membranes, e.g., protiated membranes in deuterated buffer, showing the NSLD profile along the normal to the membrane surface. The difference in the NSLD between the headgroup (~2 × 10-2 Å-2) and tail region (~4.5 × 10-6 Å-2) of the membrane is due to the headgroup hydration with deuterated buffer. Right: Tail-contrast matched membranes such that the hydrocarbon tail region of the membrane has the same NSLD as the buffer, as shown in the corresponding NSLD profile along the membrane normal. (B) Domain-forming membranes with two neutron contrast schemes where the domains (center) or the matrix (left) are contrast-matched to the buffer, enabling selective studies of matrix or domain dynamics, respectively. This figure has been modified from Nickels et al., JACS 201541. (C) Asymmetric membranes prepared by cyclodextrin exchange between protiated and deuterated lipid vesicles, resulting in the deuteration of one membrane leaflet while keeping the other leaflet protiated. This allows studies of the bending dynamics of the protiated leaflet and provides insights into the mechanical coupling between opposing leaflets in asymmetric membranes. This figure has been modified from Rickeard et al., Nanoscale 202040. Please click here to view a larger version of this figure.
to:
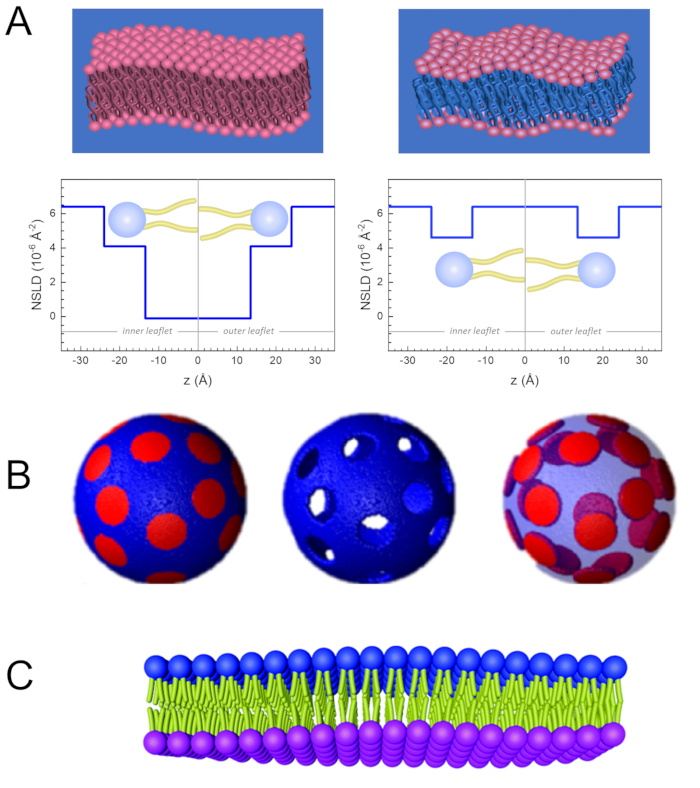
Figure 2: Examples of possible deuteration schemes in NSE experiments on lipid membranes. (A) Left: Fully contrasted membranes, e.g., protiated membranes in deuterated buffer, showing the NSLD profile along the normal to the membrane surface. The difference in the NSLD between the tail region (~0 × 10-2 Å-2) and headgroup region (~4.5 × 10-6 Å-2) of the membrane is due to the headgroup hydration with deuterated buffer. Right: Tail-contrast matched membranes such that the hydrocarbon tail region of the membrane has the same NSLD as the buffer, as shown in the corresponding NSLD profile along the membrane normal. (B) Domain-forming membranes with two neutron contrast schemes where the domains (center) or the matrix (left) are contrast-matched to the buffer, enabling selective studies of matrix or domain dynamics, respectively. This figure has been modified from Nickels et al., JACS 201541. (C) Asymmetric membranes prepared by cyclodextrin exchange between protiated and deuterated lipid vesicles, resulting in the deuteration of one membrane leaflet while keeping the other leaflet protiated. This allows studies of the bending dynamics of the protiated leaflet and provides insights into the mechanical coupling between opposing leaflets in asymmetric membranes. This figure has been modified from Rickeard et al., Nanoscale 202040. Please click here to view a larger version of this figure.
转载和许可
请求许可使用此 JoVE 文章的文本或图形
请求许可探索更多文章
This article has been published
Video Coming Soon
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。