Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Bromodeoxyuridine (BrdU) Labeling and Subsequent Fluorescence Activated Cell Sorting for Culture-independent Identification of Dissolved Organic Carbon-degrading Bacterioplankton
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
Environmental bacterioplankton are incubated with a model dissolved organic carbon (DOC) compound and a DNA labeling reagent, bromodeoxyuridine (BrdU). Afterward, DOC-degrading cells are separated from the bulk community based on their elevated BrdU incorporation using fluorescence activated cell sorting (FACS). These cells are then identified by subsequent molecular analyses.
Streszczenie
Microbes are major agents mediating the degradation of numerous dissolved organic carbon (DOC) substrates in aquatic environments. However, identification of bacterial taxa that transform specific pools of DOC in nature poses a technical challenge.
Here we describe an approach that couples bromodeoxyuridine (BrdU) incorporation, fluorescence activated cell sorting (FACS), and 16S rRNA gene-based molecular analysis that allows culture-independent identification of bacterioplankton capable of degrading a specific DOC compound in aquatic environments. Triplicate bacterioplankton microcosms are set up to receive both BrdU and a model DOC compound (DOC amendments), or only BrdU (no-addition control). BrdU substitutes the positions of thymidine in newly synthesized bacterial DNA and BrdU-labeled DNA can be readily immunodetected 1,2. Through a 24-hr incubation, bacterioplankton that are able to use the added DOC compound are expected to be selectively activated, and therefore have higher levels of BrdU incorporation (HI cells) than non-responsive cells in the DOC amendments and cells in no-addition controls (low BrdU incorporation cells, LI cells). After fluorescence immunodetection, HI cells are distinguished and physically separated from the LI cells by fluorescence activated cell sorting (FACS) 3. Sorted DOC-responsive cells (HI cells) are extracted for DNA and taxonomically identified through subsequent 16S rRNA gene-based analyses including PCR, clone library construction and sequencing.
Protokół
1. Water sample processing
- Filter 10L environmental water through 1 μm-pore-size membrane filters to remove large particles and bacteriovores. Collect the water filtrate in a carboy.
- Transfer 36 ml filtrate each into 3 sterile Eppendorf tubes (50 ml) containing 4 ml freshly prepared paraformaldehyde solution (PFA; 10%). Incubate for 2 hrs at room temperature to preserve cells. Collect cells onto 0.22-μm-pore-size white membrane filters by vacuum filtration. Wash the filter by passing 10 ml phosphate-buffered saline (PBS) through the filter by vacuum filtration. Label the filters as negative controls and store them at -20°C.
Steps 1.3-1.4 are optional for establishing DOC-limited conditions.
- Add a mixture of inorganic nitrogen and phosphorus (5 μM NH4Cl, 5 μM NaNO3, and 1μM NaH2PO4 final concentration) into the carboy.
- Incubate in the dark at in situ temperature for 48 hours with occasional agitations.
2. Microcosm establishment and incubation
- Fill each of six 1-L glass flasks with 800 ml water sample from the carboy of step 1.4 to establish microcosms. Add BrdU (10 μM, final concentration) to each microcosm. Mix well.
- Add 1 ml model DOC compound solution into three of the microcosms, these will serve as DOC amendments. Add 1 ml sterile PBS into the three remaining microcosms, these will serve as no-addition controls.
- Incubate all microcosms in an incubator shaker and incubate in the dark at in situ temperature while shaking at 100 rpm.
- Collect 36 ml of water sample from each microcosm and transfer into 50 ml sterile Eppendorf tubes at time points of 0, 8, 16, and 24 hours. Immediately add 4 ml of fresh PFA (10%) to collection tubes and incubate for 2 hrs at room temperature to preserve the cells.
- Filter preserved cells through 0.22-μm-pore-size polycarbonate membrane filters. Wash the filters with 10 ml PBS. Proceed immediately to the next step or store the filters at -20°C.
3. In situ immunodetection for BrdU Incorporation
- Thaw the filters (DOC amended samples, no-addition controls and negative controls) at room temperature.
- Apply 1 ml of lysozyme solution [10 mg/ml lysozyme egg white in 100 mM Tris, 50 nM EDTA (pH = 8)] 4 to cover bacterial cells on the filter. Incubate at room temperature for 30 minutes. Wash the filter by passing 10 ml PBS through it under suction.
- Add 1 ml of proteinase K solution [2 mg/ml proteinase K in 100 mM Tris, 50 nM EDTA (pH = 8)] 4 to cover bacterial cells on the filter. Incubate at room temperature for 30 minutes. Wash the filter by passing 10 ml PBS through it under suction.
- Add 1 ml exonuclease solution [exonuclease III (50 U/ml) in 5 mM MgCl2 and 50 mM Tris-HCl] 5 to cover bacterial cells on the filter. Incubate at 37 °C for 30 minutes. Wash the filter by passing 10 ml PBS through it under suction.
- Assemble frame-seal incubation chambers (Bio-Rad) according to the manufacturer's instructions.
- Slice the filter into eighths using a sterile blade on an alcohol-cleaned dry surface.
- Using forceps, place an eighth section of a filter into one assembled frame-seal incubation chamber (eight frame-seal chambers are needed for each filter sample). The side of the filter section that contains cells should face upwards. The moisture at the back of the filter will allow the filter stick to the slide. If filter becomes dry, apply a tiny drop (2 μl) of diH2O in the center of the chamber before placing the filter section on the slide.
Steps 3.8-3.20 use the reagents from the ROCHE In Situ Cell Proliferation Kit, FLUOS following procedures modified from the manufacturer's instructions. Except for PBS, all reagents are provided within the kit.
- Apply enough incubation buffer (0.5% bovine serum albumen, 0.1% Tween20 in PBS,) to evenly cover the entire filter section in the incubation chamber without causing overflow once the seal is applied.
- Place a polyester frame cover over the incubation chamber frame. Press down to tightly seal the incubation chamber. Avoid air bubbles above the filter. Incubate the sealed chambers for 10 min at room temperature in the dark.
- Remove the polyester seal and open the incubation chambers. (Note: opening the chambers sometimes will pull up the frame seal from the slide. In that case, prepare a new chamber for the following steps.)
- Pipet out the incubation buffer from one corner of the chamber. Avoid contact with the filter.
- Wash the filter by gently pipetting 100 μl PBS in and out from the chamber 3 times.
- Prepare anti-BrdU-FLUOS working solution following the steps recommended by the manufacturer immediately before use.
- Pipet 120 μl anti-BrdU-FLUOS working solution onto the filter, taking care to evenly cover the entire surface.
- Reseal the chamber with a new polyester cover. Avoid air bubbles on top of the filter. Incubate the chamber in the dark at 37 °C for 3 hours. This step will label BrdU incorporated DNA in situ with fluorescein isothiocyanate (FITC).
- Remove the polyester cover and open the incubation chamber. Pipet out the anti-BrdU-FLUOS working solution. Wash the filter 3 times with PBS.
- Transfer the filter from the incubation chamber to a sterile surface. Slice the filter section into small pieces using a sterile blade.
- Transfer the filter pieces into 2 ml microcentrifuge tubes, each containing 1 ml PBS. Tightly cap the tube and seal with parafilm. Incubate at 37 °C and 200 rpm for 10 min.
- Secure the tubes onto a vortexer and vortex at the maximum speed for 5 minutes. Pipet the supernatant into a sterile 15 ml Eppendorf tube. Repeat incubation and vortex steps for 5 more times. Combine the supernatant in the same 15 ml Eppendorf tube for each sample. Typically, 80% cells can be recovered in the suspension.
- Store the supernatant with resuspended cells at 4 °C. Sort within 2 days.
4. FACS analysis
A procedure for a BD FACSAria flow cytometer and corresponding software is described here.
- Optimize the setting of the flow cytometer following steps recommended by the manufacturer. This entails: adjusting the amplification control to set the required breakoff parameters and the sweet spot; optimizing the laser delay and area scaling factors for the experiment sheath pressure and optimizing settings for FSC and SSC voltages, FSC threshold, FSC fluorescence scaling, fluorescence PMT voltages, etc.
- Run negative control samples on the flow cytometer (FCM) based on fluorescence intensity of FITC and side scatter (SSC). Increase the FITC threshold until no cells in the negative controls can be visualized by the FITC-SSC acquisition display.
- Run no-addition control samples and examine the distribution pattern of 10,000 cells based on FITC and SSC acquisition. Define a gate to enclose all the cells and designate them as "low intensity cells" (LIs).
- Run DOC-amended samples and examine the distribution pattern of 10,000 cells based on FITC and SSC. Some cells will appear in the preset LI gate. Define another gate to enclose the cells that have higher fluorescence intensity (HIs) than LIs. Acquire statistics to view the relative abundance of gated cells.
- Sort HI cells into collection tubes containing 500 μl PBS at "purify 1 drop" mode. Terminate sorting when the number of HIs reaches 500,000 counts.
5. Filter PCR amplification of 16S rRNA genes
Filter PCR procedures are modified from Kirchman et al. 6
- Filter sorted cells onto a white, 0.22 μm-pore-size, 25 mm-diameter, polycarbonate membrane filter. Trim off the edge of the filter that contains no bacterial cells using a sterile blade. Slice the filter into 8 equal sized pieces.
- Place a single filter piece into a PCR reaction tube, with the cells facing inward of the tube. Add 45 μl PCR grade water into the PCR reaction tube. Submerge the filter entirely in the water.
- Add 2 ROCHE illustra PuReTaq Read-To-Go PCR Beads into the PCR reaction tube, briefly vortex. Add 2 μl each of the forward and reverse 16S rRNA gene primers (0.4 μM final concentration for each primer), such as 27F and 1492R 7, into each of the PCR reaction tubes.
- (Optional) Add 1μl bovine serum albumin solution (BSA, final concentration is 30μg/100 μl) to the PCR reaction mixture to help adsorb amplification inhibitors. If one chooses not to add BSA, add 1μl of water instead.
- Perform PCR amplification on a thermal cycler. A touch down PCR program is recommended, which has the annealing temperature sequentially decreasing from 62 to 52°C by 1°C per cycle for 11 cycles, followed by 15 cycles with annealing temperature of 52°C. All cycles include denaturing (at 95°C), annealing (at 62 to 52°C), and extension (at 72°C) steps of 50s duration. An initial 3-min denaturation and final 10-min extension step is also included in the PCR program.
- Confirm PCR amplification by electrophoresis on an ethidium bromide-stained 1% agarose gel. Excise the PCR amplicons from the gel and clean with the QIAGEN QIAquick gel extraction kit.
- Perform two additional PCR amplifications for each sample, each time use a new filter section. After PCR gel purification, pool the amplicons of the same sample together. Purified 16S rDNA amplicons are now ready for a number of molecular analyses that allow taxonomic identification, such as clone library construction and sequencing.
6. Representative results:
Representative results are described using a study of putrescine-degrading bacteria as an example. Water samples were collected from a coastal site of Georgia and processed following the procedures described above. FACS analysis revealed that putrescine addition induced development of a group of bacteria with high FITC fluorescence intensity, indicating high BrdU incorporation rate (Figure 1). These cells were designated as high-BrdU-incorporation cells (HIs) and were expected to contain mostly putrescine-degrading bacteria. HIs were missing in the no-addition controls, which only contained cells with lower levels of BrdU incorporation (LIs). LIs were expected to mainly contain bacterioplankton that were unable to use added putrescine. HIs were sorted into separated tubes and then collected onto membrane filters. 16S rRNA gene amplicons were obtained for high HI cells using filter PCR.
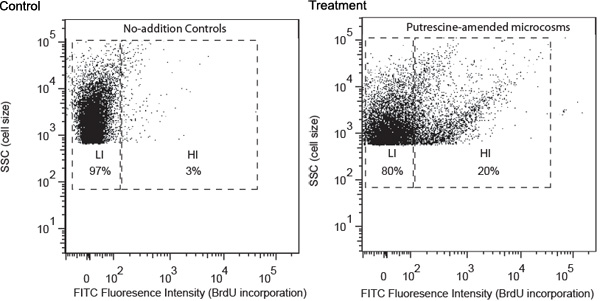
Figure 1. Flow cytometric analysis of no-addition control and model-compound-amended (putrescine as an example here) samples collected after 24 h of incubation. Cell distribution analysis was based on (1) fluorescence intensity of FITC labeling (x-axis), which is positively related to level of BrdU incorporation, and (2) side scatter (SSC, y-axis), which is positively related to cell size. Gate notation is based on level of BrdU incorporation, (HI, high-BrdU-incorporation; LI, low-BrdU-incorporation). The relative percentage of HI and LI cells are shown in corresponding gates.
Access restricted. Please log in or start a trial to view this content.
Dyskusje
Our approach couples BrdU incorporation, FACS and 16S rDNA analysis to allow species-level identification of bacterioplankton that metabolize individual DOC components in aquatic environments. The BrdU incorporation assay labels bacterial cells based on metabolic activities, which allows analysis only on active bacteria and thus does not include dormant cells. In our approach, BrdU incorporation is in situ immunodetected and bacteria that have different levels of BrdU incorporation are subsequently visualized, g...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
No conflicts of interest declared.
Podziękowania
Funding of this project was provided by the National Science Foundation grants OCE1029607 (to X.M.) and MCB0702125 (to M.A.M.) and the Gordon and Betty Moore Foundation (to M.A.M.).
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments | |
| BrdU | Reagent | Sigma-Aldrich | B5002-5G | |
| Lysozyme | Reagent | Sigma-Aldrich | L6876-5G | |
| Proteinase K | Reagent | Sigma-Aldrich | P2308-25MG | |
| In Situ Cell Proliferation Kit, FLUOS | Kit | Roche Group | 11810740001 | Consume more of Anti-BrdU-FLUOS and Incubation buffer per reaction than suggested by the manufacturer. |
| Frame-Seal Incubation Chambers | Material | Bio-Rad | SLF-1201 | |
| Polycarbonate Membrane Filters (142-mm-diameter, 1.0 μm-pore-size) | Material | EMD Millipore | FALP14250 | |
| Polycarbonate Membrane Filters (25-mm-diameter, 0.2 μm-pore-size) | Material | EMD Millipore | FGLP02500 | |
| illustra PuReTaq Ready-To-Go PCR Beads | Kit | GE Healthcare | 27-9559-01 | |
| QIAquick gel extraction kit | Kit | Qiagen | 28704 | |
| FailSafe PCR System | Kit | Epicentre Biotechnologies | FS99060 |
Odniesienia
- Pernthaler, A., Pernthaler, J., Schattenhofer, M., Amann, R. Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl. Environ. Microbiol. 68, 5728-5728 (2002).
- Urbach, E., Vergin, K. L., Giovannoni, S. J. Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl. Environ. Microbiol. 65, 1207-12 (1999).
- Mou, X. Z., Hodson, R. E., Moran, M. A. Bacterioplankton assemblages transforming dissolved organic compounds in coastal seawater. Environ. Microbiol. 9, 2025-2025 (2007).
- Hodson, R. E., Dustman, W. A., Garg, R. P., Moran, M. A. In situ PCR for visualization of microscale distribution of specific genes and gene products in prokaryotic communities. Appl. Environ. Microbiol. 61, 4074-4074 (1995).
- Dinjens, W. N. Bromodeoxyuridine (BrdU) immunocytochemistry by exonuclease III (Exo III) digestion. Histochemistry. 98, 199-199 (1992).
- Kirchman, D. L., Yu, L. Y., Fuchs, B. M., Amann, R. Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat. Microb. Ecol. 26, 13-13 (2001).
- Delong, E. F., Wickham, G. S., Pace, N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 243, 1360-1360 (1989).
- Artursson, V., Jansson, J. K. Use of bromodeoxyuridine immunocapture to identify active bacteria associated with arbuscular mycorrhizal hyphae. Appl. Environ. Microbiol. 69, 6208-6208 (2003).
- Mou, X. Z. Bacterial carbon processing by generalist species in the coastal ocean. Nature. 451, 708-708 (2008).
- Mou, X. Flow-cytometric cell sorting and subsequent molecular analyses for culture-independent identification of bacterioplankton involved in dimethylsulfoniopropionate transformations. Appl. Environ. Microbiol. 71, 1405-1405 (2005).
- Dean, F. B. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. U.S.A. 99, 5261-5261 (2002).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone