Method Article
Radioactive in situ Hybridization for Detecting Diverse Gene Expression Patterns in Tissue
W tym Artykule
Podsumowanie
This protocol is successfully used to quantitatively detect levels and spatial patterns of mRNA expression in multiple tissue types across vertebrate species. The method can detect low abundance transcripts and allows processing of hundreds of slides simultaneously. We present this protocol using expression profiling of avian embryonic brain formation as an example.
Streszczenie
Knowing the timing, level, cellular localization, and cell type that a gene is expressed in contributes to our understanding of the function of the gene. Each of these features can be accomplished with in situ hybridization to mRNAs within cells. Here we present a radioactive in situ hybridization method modified from Clayton et al. (1988)1 that has been working successfully in our lab for many years, especially for adult vertebrate brains2-5. The long complementary RNA (cRNA) probes to the target sequence allows for detection of low abundance transcripts6,7. Incorporation of radioactive nucleotides into the cRNA probes allows for further detection sensitivity of low abundance transcripts and quantitative analyses, either by light sensitive x-ray film or emulsion coated over the tissue. These detection methods provide a long-term record of target gene expression. Compared with non-radioactive probe methods, such as DIG-labeling, the radioactive probe hybridization method does not require multiple amplification steps using HRP-antibodies and/or TSA kit to detect low abundance transcripts. Therefore, this method provides a linear relation between signal intensity and targeted mRNA amounts for quantitative analysis. It allows processing 100-200 slides simultaneously. It works well for different developmental stages of embryos. Most developmental studies of gene expression use whole embryos and non-radioactive approaches8,9, in part because embryonic tissue is more fragile than adult tissue, with less cohesion between cells, making it difficult to see boundaries between cell populations with tissue sections. In contrast, our radioactive approach, due to the larger range of sensitivity, is able to obtain higher contrast in resolution of gene expression between tissue regions, making it easier to see boundaries between populations. Using this method, researchers could reveal the possible significance of a newly identified gene, and further predict the function of the gene of interest.
Protokół
1. Tissue Preparation
- Harvest fresh tissues. For avian embryos, carefully open eggs and clean the embryo in a dish with 1xPBS, two times. For adult brains, quickly remove the brain and gently wash with 1 x PBS.
- Embed embryos or adult brain into an embedded mold full of OCT tissue tek, orientating the tissue as needed for sectioning, and quickly freeze the block by placing it into a ethanol and dry ice mixture, being careful not to get the mixture inside of the block.
- Slice the frozen sample in a cryostat into 10-12 μm thick sections. For brain and embryonic tissue, the best cutting temperate is within the range of -18 °C to -20 °C.
- Mount frozen sections on super plus glass slides.
- Store the sections in a slide box at -80 °C.
2. Generation of Radioactive Riboprobes (Use radiation safety procedures of your institution)
- Generate a purified linear DNA template of your cDNA of interest either by enzyme restricted digest of a cloned fragment surrounded by RNA polymerase promoter sites from a plasmid DNA or by PCR of the insert attached to the RNA polymerase promoter binding sites. It is also possible to generate PCR fragments with the RNA polymerase promoters as part of the 3' and 5' PCR primers. Gels purify your restricted or PCR generated fragments with the GENECLEAN gel purification kit.
- To a 0.5 ml Eppendorf tube, add 0.5-1 μg of purified linear DNA template, transcription buffer, DTT, RNasin, AGC nucleotide mix solution, and RNase free water to bring the volume up to the desired amount. Then add S35-UTP and the appropriate RNA polymerase to make either antisense or sense riboprobes.
- Incubate the mixture for 1 hr in a 37 °C water bath. Add another aliquot of RNA polymerase and incubate for another 1 hr.
- Add 3M sodium acetate solution (0.1 fold of total volume) and 100% EtOH (2.5 fold of total volume) into the Eppendorf tube to precipitate the synthesized RNA riboprobe.
- Incubate in dry ice or -80 °C for 15 min or more than 3 hours to assist precipitation.
- Pellet RNA by centrifuging at 4 °C and 15,000 rpm in a table-top Eppendorf centrifuge for 30 min.
- Remove the supernatant and wash the pellet with 70% EtOH, tap with finger to mix the pellet well.
- Pellet RNA again by centrifuging at 4 °C and 15000 rpm for 30 min.
- Remove the supernatant completely by pipeting and then add 40 μl hybridization solution. Mix well by pipeting in and out.
- Put 1 μl of the solution into 3 ml Safety-Solve solution in the scintillation vial, mix well, and measure the counts in the scintillation counter.
- Place the Eppendorf with the riboprobe at -20 °C for use within one week.
3. Tissue Treatment
- In a hood, prepare fresh PBS buffered 4% paraformaldehyde solution to about 3-5 °C above room temperature. Place slides from -80 °C storage in a metal rack on dry ice, carry to working space under the hood, and then place the rack into the 4% paraformaldehyde solution, and incubate for 5 min at room temperature.
- Wash 3 times in 1 x PBS about 15 dips each (around 1 second per dip).
- In the hood, make acetylation buffer and shake it vigorously within 10 seconds. Immediately pour into a tray containing the rack of slides and incubate for 10 min, to reduce background binding of riboprobe.
- Rinse 3 times in 2 x SSPE for 15 dips.
- Dehydrate in 70%, 95% and 100% EtOH serial for 2 min in each step (does not have to be in the hood).
- Dry the slides under the hood for at least 10-15 min.
4. Hybridization
- Calculate amount of riboprobe (0.5-1 x 106 cpm per slide) and hybridization solution (100 μl per slide) needed for all slides. Prewarm the riboprobe-hybridization mix to 65 °C for 5 min to denature the riboprobe.
- Pipet 100 μl of riboprobe-hybridization solution in a line across the slide, and use a glass coverslip to spread it evenly over the tissue and coverslip. Place slides horizontally, facing upright into a metal rack and place rack slowly upright into the 65 °C oil bath for a minimum of 4 hr and a maximum of 16 hr. The oil creates an airtight seal around the coverslips.
- Remove metal racks from the oil bath and wipe the excess oil around rack with tissue paper.
- Wash off oil from the covered slides and metal rack in glass or metal trays containing chloroform, two times. The 2nd chloroform wash can be used as the first for the next experiment.
- Transfer the rack with covered slides into a tray with in 0.1% β-mercaptoethanol + 2 x SSPE solution and move up and down for a few dips to loosen coverslips and remove dissolve excess hybridization solution.
- Transfer the rack with covered slides in fresh solution of 0.1% β-mercaptoethanol + 2 x SSPE, and then remove coverslips with an RNase free forceps in solution to prevent scratching tissue sections. Transfer the uncovered slides into fresh rack in a tray horizontal slide rack holder, in a solution of 0.1% β-mercaptoethanol in 2 x SSPE.
- Incubate the rack with slides in the fresh 0.1% β-mercaptoethanol + 2 x SSPE solution at room temperature for 1 hr to remove excess unbound RNA probe. Discard this and the previous aqueous wash solutions, as well as coverslips, as radioactive waste.
- Transfer the rack with slides to a prewarmed 2x SSPE solution at 65 °C, add β-mercaptoethanol to a final of 0.1% concentration, and incubate at 65 °C for 1 hr. Discard as radioactive waste.
- Transfer and incubate the rack with slides two times in prewarmed 0.1 x SPPE at 65 °C for 30 min each. The amount of radioactivity removed in this step is very small and no longer considered excessive radioactive waste after this step.
- Dehydrate the rack and slides in 70%, 95% and 100% EtOH for 2 min each.
- Dry the slides in the hood for at least 30 min.
5. Visualization of Radioactive Signal
- Place dry slides into a film cassette and in a dark room, place the x-ray film (Kodak BioMax MR film) over the slides, and close the cassette. Make sure that the slides are facing the emulsion side of the x-ray film. Expose the slides for ~1-7 days depending on expected abundance of the transcripts.
- Develop the x-ray film in standard developer and fixer. The hybridization signal shows up as black (exposed silver grains in the emulsion) on the film (Fig. 1).
-
- (Optional) To determine cellular resolution and see signal on the tissue, the slides need to be dipped in photographic emulsion and counterstained. If you are going to eventually counterstain with cresyl violet, then delipidize sections by incubating in xylene for 5 min at room temperature twice, rehydrate 1 min each in 100%, 100%, 95%, 95%, 70%, and 50% EtOH and then in deionized water. If you are not going to stain with a dye that does not require delipidization, then delipidization is not necessary. Dry slides well under a hood for at least 2-3 hours.
- In the dark room using a safe light, scoop out enough Kodak NTB emulsion to cover half the slide lengthwise (i.e. tissue) in glass dipping container, and then melt in a 42 °C water bath for 20-30 min. Then dilute it with distilled water to 1:1 ratio. The level of the emulsion should now cover all sections on a slide when the slide is dipped in it. If you have a lot of slides, you may need to prepare extra emulsion.
- Dip slides into the diluted emulsion in the 42 °C water bath and dry dipped slides in a closed light tight container overnight in the dark room, or in an oven at 37 °C for 2-3 hours, with lights off.
- Transfer the slides into racks slots in black boxes containing desiccators, being careful for the slides not to touch each other and thus create artifacts. Seal the edges of the boxes with black electrical tape slowly to prevent static-induced light sparks and then wrap the boxes in aluminum foil. Store the boxes at 4 °C from several days to weeks (signal from 1 day on x-ray film is similar to 5 days under emulsion).
- Warm the slide boxes to room temperature for 1 hr.
- In the darkroom, remove rack with slides (or place slides in a metal rack if using boxes with slide slots) from the boxes and develop them in the Kodak D-19 developer at 16 °C for 3.5 min.
- Wash the developed slides in tap water at room temperature for 1 min.
- Incubate the slides twice in fixer at 19 °C for 6 min each. Lights can be turned on during the second fixer incubation.
- Wash the slides in running water at room temperature for at least 30 min and scrape the emulsion from the back side of the slide while 'wet' with a razor blade to prevent scratching the glass.
- Stain tissue with 0.3% cresyl violet in tap water for 5 min.
- Wash extra cresyl violet solution in fresh tap water for ~15 dips.
- Dehydrate the slides for ~15 dips in each alcohol solution: 50%, 70%, 95%, 95%, 100% and 100% EtOH.
- Incubate the slides in xylene for 5 min at room temperature twice.
- Coverslip with Permount medium on the slide and dry the covered slide in the hood overnight (>16 hr); it will take several days before the glue is sturdy enough to clean the slides further.
6. Generating Darkfield Color Images
- If necessary, further clean excess emulsion on the back of the slides (non-coversliped side without tissue) by wetting it with water and scraping with a razor blade.
- Rinse slides with 80% EtOH solution and wipe 1-2 times gently to get rid of debris and dust.
- Take pictures under darkfield or brightfield illumination. Steps 6.2 and 6.3 may have to be repeated 2-3 times in order to obtain a good images without dust particles, which are easily seen in darkfield under a dissecting microscope.
7. Representative Results
The are two main ways of viewing in situ hybridization results on tissue sections hybridized with S35 radioactive probes: 1) x-ray film that was placed over the slides or 2) emulsion that was coated on the slides. A third approach is using a phosphorimager screen placed over the slides, but we have not been satisfied with the resolution of this approach. X-ray films provide a quick result and analyses of the overall condition of the hybridization. The x-ray film data also reveals broad anatomical resolution and can be used for quantitative analysis10. Examples of x-ray film images of late avian embryo heads hybridized with antisense probes for FoxP1 and CoupTF2 gene expression are in Figures 1A and B. Both genes are highly abundant in specific brain subdivisions. A good quality x-ray film result should be sharp (not blurry) and have high signal-to-background ratio. A blurred image can be due to the uneven contact between an x-ray film and the glass slide with the hybridized tissue.
For emulsion dipped slides, the emulsion contains light sensitive silver salts coated on the tissue as apposed to being on the plastic of the x-ray film. During developing, the S35-exposed silver salts are converted to metallic silver grains, much like in the x-ray film. However, the silver deposits are directly visible over the cells representing gene expression that can be observed and measured qualitatively under a microscope. The metallic silver grains block direct light through and appear as the black dots under brightfield view. The cresyl violet counterstain appears purple in color (Fig. 3A, 3C, and Fig. 4). In darkfield, the silver grains reflect light coming from the side and appear as the white dots (Fig. 1C, 1D, 3B and 3D). In this situation, the cresyl violet stain appears red in color. In brightfield, the hybridization signal is easier to view under high magnification at cellular resolution, whereas in darkfield, in addition, the hybridization signal can be viewed under lower magnification over the entire tissue. The darkfield view is the approach we commonly use to show the overall gene expression pattern. However, relative to the quick result obtained from x-ray films, the emulsion dipped slides takes longer time (one to several weeks) and is more sensitive to obtaining background.
There are four common sources of strong background: 1) Background all over the x-ray film is usually do to problems with the developer or fixer, or partially exposed film; 2) Background on the glass slides is usually due to problems with washing or glass slide preparation, such as improper silination of the slides from the commercial source or self-prepared; 3) Emulsion exposure and development background; and 4) Background on the section due either to lack of careful post-hybridization wash steps, too low of a hybridization temperature, poor quality of the hybridization solution, paraformaldehyde contamination in washing dishes causing probes to permanently cross-link to the tissue, riboprobe degradation leading to small molecules labeling the tissue non-specifically, inactive DTT or β-mercaptoethanol resulting in cross linking of S35-RNA probes in di-sulfide bonds to the tissue, and waiting too long for acetylation. It is critical to have the slides in the acetylation solution within seconds of mixing the acetic anhydride and triethanolamine. If several minutes pass without adding the solution to the slides, then acetyl groups will not be efficiently removed and then bind to RNA non-specifically. Other factors include hybridization over 20 hr, which may generate too strong of a signal, and excess oil droplets on the slides, which sequester hybridization solution on the slides during aqueous washes, resulting in radioactive spots on the tissue and slides giving dark background signals. If working with many slides (more than 100 slices), add a 3rd chloroform wash or change the chloroform washes, to prevent excess oil particles from remaining on the slides. Careless tissue treatment, i.e. not freezing quickly enough (within 5-10 min after dissection) or thawing and re-freezing also increases background due to mRNA degradation. Be careful not to mistake background for over exposure.
For emulsion background on the dipped slides is possible because it is highly light sensitive and requires long-exposure in the dark. Common background problems include too high of temperature for the developer and fixer. When the temperature is higher than 19 °C, close to or warmer than room temperature, more silver grain background is obtained. Exposure to low levels of light leaking into a darkroom will cause emulsion background. Not washing out fixer long enough (at least 30 min in running water), will leave fixer that then reacts with cresyl violet to generate a brownish precipitate throughout the emulsion. However, if the slides are washed in water longer than 90 min after fixation before cresyl violet staining, this can cause the emulsion to become loose and the sections to stain poorly. If there is not enough time to stain the slides within a 30-90 min window after fixation and washing, after the 30 min wash, dry the slides overnight and proceed with cresyl violet staining the next day. Generally, most mRNAs have specific gene expression patterns, whereas background signal is more uniform.
Folded tissue could be misleading for gene expression results on x-ray film, leading to a region with darker signal. To determine if the tissue is folded, examine non-stained sections under darkfield or cresyl violet stained sections under brightfield. Cresyl violet counterstaining provides a better way to examine the tissue condition.
For a better interpretation of probe specificity, control sense probes should be applied on several adjacent sections. Most sense probes do not show a signal, but some do, and when they do, we find that it is often different than the antisense signal. We believe that this could be related to antisense synthesis or another gene on the antisense strand of the genome. We present an example Pax6 sense and antisense probes (Fig. 2A and B). The antisense strand reveals labeling along the ventricular zone of the forebrain, cerebellum and eye as expected (Fig. 2A), but the sense reveals labeling in the pigment layer of retina (Fig. 2B).
For probe sizes, we use cDNA probes anywhere in the range of 300-5000 bps. Probes less than 300 bps work, but the signals are usually weaker. We have not tried probes bigger than 5000 bps. It is best to use probes on tissue from the same species if possible. If not, we cross-hybridize probes on sections of other species and decrease the hybridization and wash temperatures in 3-5 °C increments based on sequence identity, if known. If not known, then we perform trial and error hybridization and wash temperatures. If the temperature is lowered too much, the cDNA probe can cross-hybridize with other mRNAs of similar sequences across species or in the same species11. In practice, we find that cDNAs that are ~95% identical or greater to the mRNA target in the tissue, the stringent hybridization and wash temperature (65 °C) conditions works well. For sequences that are in the ranges of ~85 to 94% identical, the hybridization and wash temperature may need to be reduced in the range of ~50 to 60 °C.
The reason for using a metal rack in most of the steps is the use of chloroform washes and xylene. Both organics melt many kinds of plastics. Glass and some kinds of plastics are resistant to these organics. But glass is easier to break, and some plastics that at first are resistant will melt over long-periods of exposure to the organics.
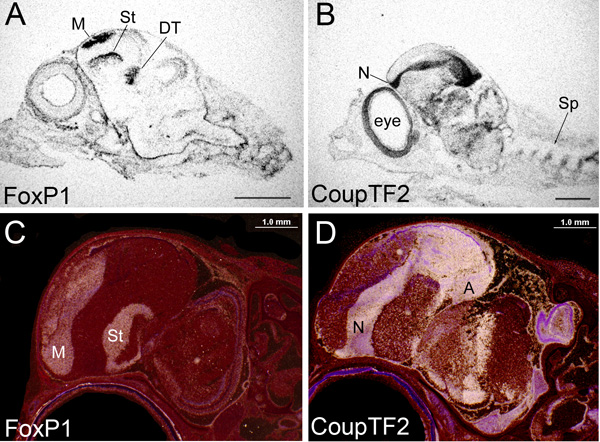
Figure 1. Autoradiography of in-situ hybridization images from x-ray films and emulsion dipped slides. (A-B) X-ray film images of sagittal whole head sections of the zebra finch songbird at embryonic day 10, hybridized with antisense riboprobes to (A) FoxP1 or (B) CoupTF2, taken with brightfield illumination under a dissecting microscope. Black, exposed grains in film showing mRNA expression. Scale bar = 500 μm. (C-D) Emulsion dipped slide images of sagittal whole head sections of the zebra finch at post hatch day 6, hybridized with antisense riboprobes to (C) FoxP1 and (D) CoupTF2, taken with darkfield illumination under a dissection microscope. White, exposed silver grains in emulsion above tissue showing mRNA expression. Red, cresyl violet stain. Scale bar = 200 μm. For all picture, the beak is rostral to the left. The x-ray film images were exposed for one day, dipped slides for 3 days. The FoxP1 probe is 178 bps to the 1544-1711 bp part of the mRNA; CoupTF2 is 545 bps to the 1-545 bp part of the mRNA. As can be seen, within the forebrain FoxP1 mRNA is enriched in the mesopallium (M), striatum (St) and dorsal thalamus (DT), whereas CoupTF2 is enriched in the nidopallium (N), arcopallium (A), and more ventral thalamus. There is consistency in expression between exposure types (and ages). With the dipped slides, however, a higher resolution labeling is seen, and tissue boundaries and subdivisions are directly identified. These and all other images shown in the paper are from sections using the standard 65 °C high stringency hybridization.
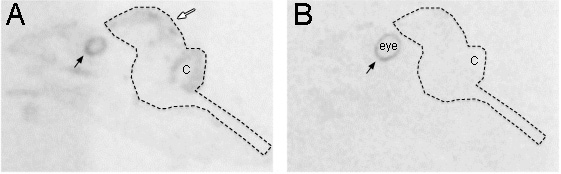
Figure 2. Comparison of antisense and sense labeling that shows different patterns. (A) The antisense strand of Pax6 was expressed in the brain, especially the ventricular zone (white arrow). Shown is autoradiography on x-ray films of sagittal whole head slices taken from zebra finch at embryonic day 12. (B) Adjacent section hybridized with the sense strand of Pax6 reveals no background expression throughout the embryo head, but apparent expression in the pigment layer of retina (black arrows) as the antisense strand. The dashed line indicates the contour of the whole brain. C: Cerebellum.
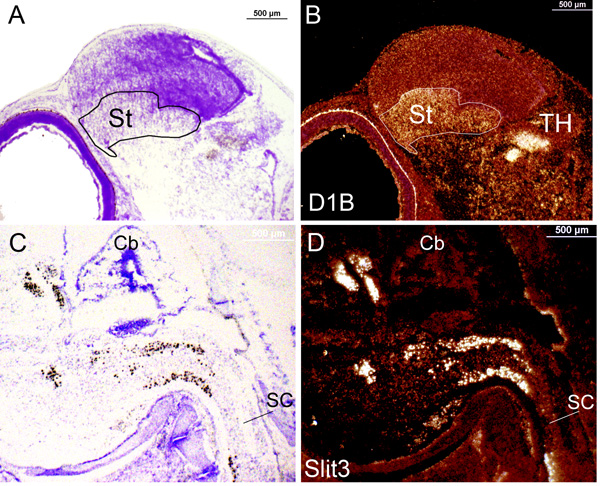
Figure 3. In situ signals of gene expression in emulsion dipped slides taken from zebra finch brain during late embryonic stages under brightfield and darkfield views. (A) Brightfield image of D1B expression at embryonic day 10 from a normal exposure to the emulsion. The label (black) can barely be seen at this magnification. Probe is 625 bps to the 1-625 bp part of the mRNA. (B) Identical section and magnification as in (A) but switched to the darkfield view showing label (white) in the striatum (St) and thalamus (TH). (C) Brightfield image of Slit3 expression at embryonic day 12 from an over exposure to the emulsion. Label (black) can easily be seen. Probe is 779 bps to the 1243-2021 part of the mRNA. (B) Identical section and magnification as in (A) switched to darkfield view showing label (white) in the spinal cord (SC) that matches the brightfield image. Rostral is oriented to the left. Cb: Cerebellum.

Figure 4. Silver grain resolution at the cellular level. Shown is FoxP1 mRNA label in the zebra finch forebrain with silver grains (black spots) above cells (cresyl violet) in different brain regions and ages compared with lower power images of Figure 1A and 1C. (A) High abundance expression over individual cells (black arrows) in the adult zebra finch mesopallium. (B) Low abundance expression over individual cells (black arrowheads) in the adjacent nidopallium of the same section. (C) High FoxP1 mRNA expression over cells (black arrows) in the embryonic day 12 mesopallium. (D) Low abundance expression over cells (black arrowheads) in the adjacent nidopallium of the same section. Example cells are circled with a yellow line. Embryonic cells (C and D) are smaller and more tightly packed compared with the adult cells (A and B). The since there is some space between the cells and emulsion, the area of exposed silver grains from the S35 probe are slightly large than the area of the cell bodies. Scale bar = 10 μm.
Dyskusje
Radioactive in situ hybridization of mRNA expression is widely used for multiple purposes, including for studying regional tissue organization, cell types, and brain functional activity2-5,10,12-14. The later use is on genes whose mRNA expression in the brain is dependent on increased neural activity, often called activity-dependent genes or immediate early genes. With these uses, our method has been applied across multiple species, including in birds, mammals (e.g. human), fish, and amphibians; in multiple tissues, including brain, skin, and muscle; and multiple ages, including hatchlings/neonates, juveniles, adults, and here in whole embryo sections2,3,5,15-17. The special features of our protocol include: (1) It produces a balance between anatomical specificity and quantitative specificity. To quantify the gene expression on the x-ray film, we take digital pictures of the images (ex: Fig. 1A and 1B), use the Photoshop (Adobe) histogram function to measure the pixel density in the regions of interest and subtract the background levels on the film outside of the tissue but still on the glass slide2,4. To quantify expression at the cellular level, we take images of silver grains over cells under high magnification (40-100X; Fig 4). We then use the threshold and measure functions of Image J by Wayne Rasband at NIH to count the number of silver grains in the image, subtract out the background count in a similar area without cells on the glass slide, divide by the number of cells, to obtain a values of expression per cell.4,18 (2) It can be relatively high throughput, allowing processing of 100-200 slides simultaneously, due to the tight coverslip seal created by the mineral oil bath. Standard in-situ hybridization methods take longer to seal the slides with parafilm, nail polish, and other means, where slides take up a lot of space; (3) It is highly sensitive for low abundance transcripts due to Dextran sulfate and Denhardt's solution in the hybridization buffer2,13; (4) The imaging approach in darkfield yields high contrast images due to taking pictures under darkfield illumination on a dissecting microscope4,5. Additionally, It allows sensitive detection of small changes in gene expression, such as in activity-dependent gene expression to identify specific brain regions activated during perception and production of specific behaviors19. The limitations relative to non-radioactive protocols are that the latter are clearer in the cellular resolution and location of the mRNA within the cell, and working with emulsion is very sensitive to manipulation and light. It is possible to combine our method with other methods, such as non-radioactive in situ hybridization to label mRNA expression of more than one gene in the same tissue10,17,20. It can be combined with immunocytochemistry to label both RNA and protein expression on the same sample in order to co-localize the mRNA with certain cell types10. The protocol modifications necessary for such double labeling experiments are described in the cited references.
In summary, our approach facilitates understanding of the timing and cellular location of gene expression in order to understanding region organization, tissue functional activity, and gene function.
Ujawnienia
We have nothing to disclose.
Podziękowania
The authors would like to thank all Jarvis lab members who improved the protocol over the years.
Materiały
| Name | Company | Catalog Number | Comments |
| Name of the reagent | Company | Catalogue number | Comments |
| Acetic anhydride | VWR | MK242002 | |
| Chloroform | VWR | BDH1109 | |
| Cresyl violet acetate | Sigma | C5042 | |
| Cryostat | Thermo scientific | Microm HM550 | |
| Deionized formamide | Sigma | F9037 | |
| DTT | Promega | P1171 | 100mM |
| EDTA | Sigma | ED | |
| Embedding mold | VWR | 15160-215 | |
| Fisher brand Superfrost plus slide | Fisher Scientific | 22-034-979 | |
| Formaldehyde | VWR | BDH0506-4LP | |
| Formamide | Sigma | F7508 | |
| GENECLEAN kit | Q-Bio gene | 1001-200 | |
| Kodak BioMax MR film | Sigma | Z350370 | |
| Kodak NTB Emulsion | Carestream Health | 8895666 | |
| Kodak Professional developer D19 | Kodak | 1462593 | |
| Kodak professional Fixer | Kodak | 1971746 | |
| β-mercaptoethanol | Calbiochem | 444203 | |
| Mineral oil | VWR | IC15169491 | |
| NaOH | VWR | SX0600-1 | |
| Paraformaldehyde | Sigma | 76240 | |
| Poly A | Invitrogen | POLYA.GF | |
| rATP | Promega | P1132 | 10mM |
| rCTP | Promega | P1142 | 10mM |
| rGTP | Promega | P1152 | 10mM |
| RNasin | Promega | N2111 | 40Units/μl |
| S35 UTP | PerkinElmer | NEG039C001MC | |
| Safety-Solve solution | Safety Solve Research Products International | 111177 | |
| Sodium Acetate | Sigma | S7899 | 3M |
| Sodium phosphate dibasic | Sigma | S3264 | |
| Sodium phosphate monobasic | Sigma | S3139 | |
| SP6 RNA polymerase | Promega | P1085 | |
| Staining metal rack | Electron Microscopy Sciences | 70312-54 | |
| T7 RNA polymerase | Promega | P2075 | |
| Tissue-Tek OCT | Sakura | 4583 | |
| 5x Transcription buffer | Promega | P1181 | |
| Triethanolamine | VWR | IC15216391 | |
| Tris-HCl (1 M, pH 8.0) | VWR | 101449-446 | |
| tRNA | Roche | 10109509001 |
Solutions:
- Reagents for making of riboprobes: 1.5 μl DNA template (0.2-0.3 μg/μl), 2 μl of 5x optimized transcription buffer, 1 μl of 100mM DTT (comes with Polymerase from Promega, mix well at room temperature), 0.3 μl of RNasin (40 units/μl), 1.5 μl of AGC ribonucleotide mix solution, 3.5 μl of S35 UTP, and 1 μl RNA polymerase. Bring up to 10 μl with nuclease-free water.
- AGC ribonucleotide mix solution: mix equal amounts of 10mM ATP, GTP and CTP together.
- Sodium acetate solution for EtOH precipitation to remove free S35 UTP: 40 μl RNase and DNase free water, 5 μl of 3M Sodium Acetate, and 125 μl of 100% EtOH.
- Hybridization buffer (10ml stock): 5 ml of 100% deionized formamide, 600 μl of 5M NaCl, 1M tris-HCl (pH = 8.0), 240 μl of 0.5M EDTA (pH =8.0), 100 μl of 100x Denhart's solution, 100 μl of 1M DTT, 250 μl of 20mg/ml tRNA, 125 μl of 20 mg/ml poly A, and 1 g of Sodium Dextran Sulfate. Add DEPC-treated water to bring the total volume to 10 ml. Shake vigorously and then incubate at 55°C until all sodium dextran sulfate is dissolved. Store the hybridization buffer in -20°C, which is good for ~6 months.
- 4% buffered paraformaldehyde solution: Add 40 g of paraformaldehyde in 760 ml distilled water in a flask designated for paraformaldehyde, heat to 50°C on a hot plate while stirring. Add 320 μl of 10N NaOH to help dissolve the paraformaldehyde. After dissolving (~10 min), add 100 ml of 10x PBS, and bring the volume up with distilled water until 1 liter. Stir and heat the solution until paraformaldehyde is dissolved. The pH should be 7.4.
- Acetylation buffer: 13.6 ml of triethanolamine plus 2.52 ml of acetic anhydride in 1 liter of distilled water.
- 20x SSPE solution: 3M NaCl, 200 mM NaH2PO4-H2O, and 200 mM EDTA in distilled water. Adjust solution to pH 7.4 with 10N NaOH.
- 10x PBS buffer: 80g of NaCl, 2.0g of KCl, 14.4g of Na2HPO4, and 2.4g of KH2PO4 in distilled water. Adjust solution to pH 7.0 and bring total volume to 1 liter.
- Second wash solution: 0.1% β-mercaptoethanol and 50% formamide in 2x SSPE
- Cresyl violet acetate solution: 3% cresyl violet acetate in tap water (distilled water prevents good staining), dissolved overnight with stirring in a flask at room temperature. Filter with vacuum suction through a 1mm Whatman filter paper and Buchner funnel.
Odniesienia
- Clayton, D. F., Huecas, M. E., Sinclair-Thompson, E. Y., Nastiuk, K. L., Nottebohm, F. Probes for rare mRNAs reveal distributed cell subsets in canary brain. Neuron. 1, 249-261 (1988).
- Wada, K., Sakaguchi, H., Jarvis, E. D., Hagiwara, M. Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J. Comp. Neurol. 476, 44-64 (2004).
- Haesler, S. FoxP2 expression in avian vocal learners and non-learners. J. Neurosci. 24, 3164-3175 (2004).
- Jarvis, E. D., Nottebohm, F. Motor-driven gene expression. Proc. Natl. Acad. Sci. U.S.A. 94, 4097-4102 (1997).
- Holzenberger, M. Selective expression of insulin-like growth factor II in the songbird brain. J. Neurosci. 17, 6974-6987 (1997).
- Mahmood, R., Mason, I. In-situ hybridization of radioactive riboprobes to RNA in tissue sections. Methods Mol. Biol. 461, 675-686 (2008).
- Wilkinson, D. G., Nieto, M. A. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 225, 361-373 (1993).
- Acloque, H., Wilkinson, D. G., Nieto, M. A. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods Cell Biol. 87, 169-185 (2008).
- Moorman, A. F., Houweling, A. C., de Boer, P. A., Christoffels, V. M. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J. Histochem. Cytochem. 49, 1-8 (2001).
- Horita, H., Wada, K., Rivas, M. V., Hara, E., Jarvis, E. D. The dusp1 immediate early gene is regulated by natural stimuli predominantly in sensory input neurons. J. Comp. Neurol. 518, 2873-2901 (2010).
- Braissant, O., Wahli, W. A simplified in situ hybridization protocol using non-radioactively labelled probes to detect abundant and rare mRNAs on tissue sections. Biochemica. 1, 10-16 (1998).
- Kubikova, L., Wada, K., Jarvis, E. D. Dopamine receptors in a songbird brain. J. Comp. Neurol. 518, 741-769 (2010).
- Wada, K. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc. Natl. Acad. Sci. U.S.A. 103, 15212-15217 (2006).
- Jarvis, E. D. Avian brains and a new understanding of vertebrate brain evolution. Nat. Rev. Neurosci. 6, 151-159 (2005).
- Hoke, K. L., Ryan, M. J., Wilczynski, W. Social cues shift functional connectivity in the hypothalamus. Proc. Natl. Acad. Sci. U.S.A. 102, 10712-10717 (2005).
- Burmeister, S. S., Jarvis, E. D., Fernald, R. D. Rapid behavioral and genomic responses to social opportunity. PLoS. Biol. 3, e363 (2005).
- Jarvis, E. D., Schwabl, H., Ribeiro, S., Mello, C. V. Brain gene regulation by territorial singing behavior in freely ranging songbirds. Neuroreport. 8, 2073-2077 (1997).
- Jarvis, E. D., Scharff, C., Grossman, M. R., Ramos, J. A., Nottebohm, F. For whom the bird sings: context-dependent gene expression. Neuron. 21, 775-788 (1998).
- Feenders, G. Molecular mapping of movement-associated areas in the avian brain: a motor theory for vocal learning origin. PLoS One. 3, e1768 (2008).
- Chen, C. C., Fernald, R. D. Distributions of two gonadotropin-releasing hormone receptor types in a cichlid fish suggest functional specialization. J. Comp. Neurol. 495, 314-323 (2006).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone