Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Single Oocyte Bisulfite Mutagenesis
W tym Artykule
Podsumowanie
Bisulfite mutagenesis is the gold standard for analyzing DNA methylation. Our modified protocol allows for DNA methylation analysis at the single-cell level and was specifically designed for individual oocytes. It can also be used for cleavage-stage embryos.
Streszczenie
Epigenetics encompasses all heritable and reversible modifications to chromatin that alter gene accessibility, and thus are the primary mechanisms for regulating gene transcription1. DNA methylation is an epigenetic modification that acts predominantly as a repressive mark. Through the covalent addition of a methyl group onto cytosines in CpG dinucleotides, it can recruit additional repressive proteins and histone modifications to initiate processes involved in condensing chromatin and silencing genes2. DNA methylation is essential for normal development as it plays a critical role in developmental programming, cell differentiation, repression of retroviral elements, X-chromosome inactivation and genomic imprinting.
One of the most powerful methods for DNA methylation analysis is bisulfite mutagenesis. Sodium bisulfite is a DNA mutagen that deaminates cytosines into uracils. Following PCR amplification and sequencing, these conversion events are detected as thymines. Methylated cytosines are protected from deamination and thus remain as cytosines, enabling identification of DNA methylation at the individual nucleotide level3. Development of the bisulfite mutagenesis assay has advanced from those originally reported4-6 towards ones that are more sensitive and reproducible7. One key advancement was embedding smaller amounts of DNA in an agarose bead, thereby protecting DNA from the harsh bisulfite treatment8. This enabled methylation analysis to be performed on pools of oocytes and blastocyst-stage embryos9. The most sophisticated bisulfite mutagenesis protocol to date is for individual blastocyst-stage embryos10. However, since blastocysts have on average 64 cells (containing 120-720 pg of genomic DNA), this method is not efficacious for methylation studies on individual oocytes or cleavage-stage embryos.
Taking clues from agarose embedding of minute DNA amounts including oocytes11, here we present a method whereby oocytes are directly embedded in an agarose and lysis solution bead immediately following retrieval and removal of the zona pellucida from the oocyte. This enables us to bypass the two main challenges of single oocyte bisulfite mutagenesis: protecting a minute amount of DNA from degradation, and subsequent loss during the numerous protocol steps. Importantly, as data are obtained from single oocytes, the issue of PCR bias within pools is eliminated. Furthermore, inadvertent cumulus cell contamination is detectable by this method since any sample with more than one methylation pattern may be excluded from analysis12. This protocol provides an improved method for successful and reproducible analyses of DNA methylation at the single-cell level and is ideally suited for individual oocytes as well as cleavage-stage embryos.
Protokół
DAY 1
Prepare the following solutions fresh on the day of oocyte collection with sterile, distilled water such as GIBCO water. To reduce the chance of DNA contamination, change gloves often and use filter tips. Keep tubes angled away when open, and recap all tubes when not in use. We recommend that solutions are made as n+1.
3% LMP Agarose
30 mg Low Melting Point (LMP) Agarose
up to 1 ml GIBCO H2O
dissolve @ 70 °C
Lysis Solution
8 μl lysis buffer
1 μl proteinase K
1 μl 10% IGEPAL
place on ice until ready for use
2:1 Agarose:Lysis Solution (10 μl per individual oocyte, amount is for 3 oocytes)
20 μl 3% LMP agarose
10 μl Lysis Solution
mix @ 70 °C
SDS Lysis Buffer (501 μl per individual oocyte)
| 1x TE pH 7.5 | 450 μl |
| 10% SDS | 50 μl |
| Proteinase K | 1 μl |
| 501 μl |
1. Oocyte Collection
- Place the dissected mouse oviducts in M2 media, and tear the ampullae to extract the cumulus cell complex.
- Separate the oocytes from the cumulus cell complex using 0.3 mg/ml hyaluronidase solution in a 30 μl drop of M2 media. Keep the oocytes in solution only as long as it takes to remove the cumulus cells, as lengthy exposure may damage them. Wash the oocytes 3x in 30 μl drop of M2 media, removing cumulus cells periodically.
- Remove the zona pellucida using acidic tyrode's solution. Place the oocytes in one 30 μl drop of solution first, and then transfer to another 30 μl drop, as any media carried along will dilute the acid and reduce its efficiency. Keep the oocytes in solution only as long as it takes to remove the zona, as lengthy exposure may damage them. Note: an increased concentration of acidic tyrode's solution or pronase may be used for human samples, as the human zona pellucida is more resistant to treatment with acidic tyrode's solution than the mouse.
- Wash the oocytes once more in a 30 μl drop of M2 media.
2. Agarose Embedding and Lysis
- To perform agarose embedding, place the lysis solution on a 70 °C heatblock. Add the preheated LMP agarose to the lysis solution, producing a 2:1 agarose:lysis solution.
- Place a single oocyte onto a clean glass slide in minimal M2 media. Take up 10 μl of the agarose:lysis solution into a pipette tip, and (under a microscope) gently expel a small amount (~1 μl or less) onto the glass slide, allowing it to mix with the minimal media. Gently pick up the oocyte into the pipette tip and put all 10 μl into an Eppendorf tube with 300 μl mineral oil so the bead forms a sphere.
Note: this process must be done fairly quickly as the agarose will harden if the temperature drops as little as 5 °C below 70 °C. - Incubate the tube on ice for 10 minutes. To perform lysis, remove the 300 μl mineral oil and add 500 μl of the SDS lysis buffer. Incubate overnight in a 50 °C water bath.
Note: Lysis solution (Table 1) may also be used for this purpose.
DAY 2
Prepare the following solutions fresh on the day of bisulfite mutagenesis. To reduce chance of DNA contamination, change gloves often and use filter tips. Keep tubes angled away when open, and recap all tubes when not in use. We recommend that solutions are made as n+1.
| 3 M NaOH | 2.4 g NaOH in 20 ml autoclaved ddH2O |
| 0.1 M NaOH | 0.5 ml of 3M in 14.5 ml autoclaved ddH2O |
| 0.3 M NaOH | 1.5 ml of 3M in 13.5 ml autoclaved ddH2O |
2.5 M Bisulfite Solution
- 3.8 g sodium bisulfite
5.5 ml GIBCO distilled H2O
1 ml 3 M NaOH
dissolve @ room temperature - 110 mg Hydroquinone
1 ml GIBCO distilled H2O
dissolve @ 90 °C (for only as long as it takes to dissolve, mix regularly)
When fully dissolved, mix solution (a) and (b)
*Keep away from light*
3. Bisulfite Mutagenesis
- Fully remove the 500 μl SDS lysis buffer and add 300 μl mineral oil (~20 hours). Any lysis buffer remaining will dilute the agarose when it is heated and the bead will be more susceptible to dissolving in the subsequent steps. Proceed with bisulfite mutagenesis immediately, or store at -20 °C for up to 5 days.
- If applicable, remove oocytes from the freezer and let thaw (only until agarose bead is relatively translucent). Incubate for 2.5 minutes on a 90 °C heat block, following which Incubate on ice for 10 minutes.
Note: Do not mix or stir, extend longer than 2.5 minutes, or fluctuate temperature. - To perform denaturation, remove the mineral oil and add 1 ml 0.1M NaOH to each tube, flick and invert 5-6 times.
- Incubate for 15 minutes in a 37 °C waterbath, inverting every 3-4 minutes. The bead should float in the NaOH.
- To perform bisulfite treatment, spin the tube gently, then remove the NaOH and add 300 μl mineral oil and 500 μl bisulfite solution. Incubate the tube for 3.5 hours in a 50 °C waterbath. *Keep away from light*
Note: Length of incubation may need to be empirically determined for gene of interest. - To perform desulfonation, incubate on ice for 3 minutes, then remove the mineral oil and the bisulfite solution, spin gently, and add 1 ml 0.3 M NaOH. Flick and invert 5-6 times.
- Incubate for 15 minutes in a 37 °C waterbath, inverting every 3-4 minutes. The bead should float in the NaOH.
- Wash the samples, by first spinning gently, then remove the NaOH and add 1 ml 1x TE pH 7.5. Shake for 5-10 minutes at room temperature (on a shaker). Spin gently again, then remove the 1x TE. Repeat this washing process twice.
- Add 1 ml autoclaved ddH2O. Shake for 5-10 minutes at room temperature (on a shaker). Spin gently, then remove the H2O. Repeat ddH2O wash twice.
- Check the pH of the supernatant; it should be pH 5.0. If still too basic, wash again with H2O. Remove all supernatant, leaving only the agarose bead.
4. 1st and 2nd Round PCR amplification
- Prepare 1st round PCR mix **while washing**
| 10 μM Primer Forward Outer | 0.5 μl |
| 10 μM Primer Reverse Outer | 0.5 μl |
| 240 ng/ml tRNA | 1 μl |
| H2O | 13 μl |
Add to Illustra Ready-to-Go Hot Start PCR beads
Carefully slide the solid agarose bead into the PCR tube (~10 μl)
Heat to 70 °C and mix
Add 25 μl mineral oil
Total: 50 μl
- Amplify
Note: An example of cycling conditions for mouse Snrpn is denaturation for 2 minutes at 94 °C, followed by 40 cycles of 30 seconds at 94 °C, 1 minute at 50 °C, and 1 minute at 68 °C; and a final 10 minute elongation step at 68 °C. Annealing temperature for 1st round PCR for mouse H19 and Peg3 is 50 °C. - Prepare 2nd round PCR mix
| 10 μM Primer Forward Inner | 0.5 μl |
| 10 μM Primer Reverse Inner | 0.5 μl |
| H2O | 19 μl |
Add to Illustra Ready-to-Go Hot Start PCR beads
Add 5 μl 1st Round product as a template. Heat the 1st round product to 70 °C for 1 minute to soften the agarose. Be sure to pipette below the layer of mineral oil.
Add 25 μl mineral oil
Total: 50 μl
Note: Nested primer sequences for Snrpn, H19, and Peg3 can be found in Market-Velker et al10,12.
- Amplify
Note: Cycling conditions for mouse Snrpn is denaturation for 2 minutes at 94 °C, followed by 40 cycles of 30 seconds at 94 °C, 1 minute at 50 °C, and 1 minute at 68 °C; and a final 10 minute elongation step at 68 °C10. Mouse H19 and Peg3 require a 50 °C annealing temperature for 2nd round PCR. - As a diagnostic test, second round samples can be cut with a restriction enzyme that is methylation- or strain-specific.
| 2nd Round product | 4 μl |
| Restriction Enzyme | 1 μl |
| Buffer | 1 μl |
| H2O | 4 μl |
- Electrophorese the digestion products on an 8% acrylamide gel. Any heterogeneous bands represent more than one sequence.
5. TA Cloning and Colony PCR
- To clone 2nd round product, first heat to 70 °C for 1 minute to soften the agarose, then ligate into vector using the Promega pGEM-T Vector System (Fisher Scientific Cat#A1360).
| 2nd Round PCR | 1 μl |
| pGEMT-EASY vector | 1 μl |
| Ligase | 1 μl |
| H2O | 2 μl |
| 2x Ligation Buffer | 5 μl |
Incubate overnight @ 4 °C in PCR machine.
- Thaw competent E.coli cells on ice for 15 minutes (Zymo Research Corp Cat#T3009). Add 3 μl ligation reaction to 8 μl E.coli and incubate ligation on ice for 15 minutes.
- Heat shock for 40 seconds in a 42 °C waterbath, and incubate on ice for 2 minutes. Add 60 μl S.O.C. medium and incubate at 37 °C for 1 hour (in shaker).
- Place all of the reaction mix on an LB/Agar/IPTG/Xgal/Amp plate and incubate plate at 37 °C overnight.
- Prepare colony PCR mix
| 20 μM M13 Forward Primer | 0.7 μl |
| 20 μM M13 Reverse Primer | 0.7 μl |
| 5X Green Go Taq Buffer | 7.0 μl |
| 10 mM dNTP | 0.7 μl |
| Taq DNA polymerase | 0.28 μl |
| H2O | 25.62 μl |
| 35 μl Total |
Add 35 μl Colony PCR master mix into a PCR tube. Pick a white bacterial colony from the plate with a pipette tip, and swirl it into the PCR reaction.
- Amplify with denaturation for 10 minutes at 94 °C, followed by 30 cycles of 45 seconds at 94 °C, 30 seconds at 57 °C, and 1 minute at 72 °C; and a final 10 minute elongation step at 72 °C. Electrophorese 4 μl on a 1.5% agarose gel. Send ~30 μl of the PCR product for sequencing.
Note: For oocytes, 5 colony PCR products are sequenced. - Once sequencing results are obtained, methylation patterns can be read. Any original CG that remained as a CG was methylated, and any original CG that is now a TG was unmethylated.
6. Representative Results
In our work, we assay imprinted methylation in individual oocytes and embryos (Figure 1). Following nested PCR amplification using bisulfite converted primers, it is possible to confirm a successful conversion by visualizing a correct fragment size on an agarose gel (Figure 2). An individual oocyte represents one parental allele, and in theory, has one imprinted methylation pattern. As such, second round PCR products can be tested for unintentional contamination. A restriction enzyme sensitive to DNA methylation (such as HinfI or DpnII) can be used to digest the second round PCR product to assess whether it contains a methylated or unmethylated allele (Figure 3). A methylated C within the enzyme recognition sequence is cleaved while an unmethylated C that is converted to T is no longer recognized by the enzyme and is uncut. Any MII oocyte sample containing both methylated and unmethylated alleles should be discarded, as it is indicative of cumulus cell contamination (Figure 3). Following ligation and transformation, successful colony PCR amplification can be visualized on an agarose gel to ensure samples with the correct product size are sent for sequencing (Figure 4). Finally, the sequence of five individual clones from an MII oocyte should produce five identical methylation patterns and identical nonCpG conversion rates (Figure 5a). Any samples that contain more than one pattern should be discarded (Figure 5b). Since ovulated MII oocytes have two chromosome copies or an attached polar body, there is a possibility for obtaining two similar sequence patterns (Figure 5c). We recommend discarding data from oocytes that have highly dissimilar methylation patterns since cumulus cell contamination cannot be ruled out.

Figure 1. Schematic of the Single Oocyte Bisulfite Mutagenesis assay.
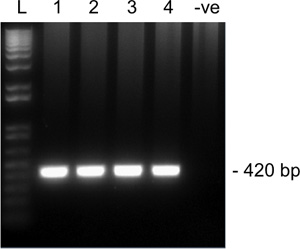
Figure 2. Representative results from 2nd round amplification for Snrpn from a single MII oocyte on a 1.5% agarose gel. Lanes 1-4 are four single MII oocytes and lane 5 is a negative control (no oocyte). Expected amplicon size for Snrpn is 420 bp. L, ladder.
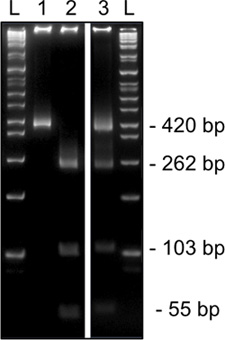
Figure 3. Representative results from 2nd round methylation-specific restriction digestion for Snrpn from a single MII oocyte on an 8% acrylamide gel. HinfI diagnostic restriction digestion shows unmethylated DNA which harbors a T that abolishes the restriction site (420bp, lane 1) or methylated DNA which contains a C within recognition site (cut, 262, 103, and 54 bp, lane 2). Digestion showing both methylated and unmethylated restriction enzyme sites (cut & uncut bands, lane 3) are indicative of cumulus cell contamination. L, ladder.
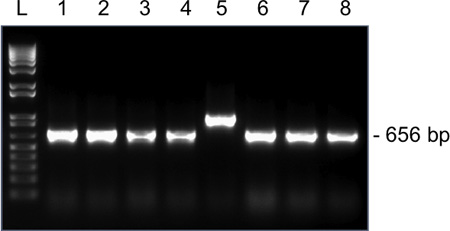
Figure 4. Representative results for colony PCR amplification for Snrpn from a single MII oocyte on a 1.5% agarose gel. Expected amplicon size following ligation of Snrpn into the pGEM-T Easy vector and using M13 forward and reverse primers is 656 bp. Lane 1-8, amplicons from clones 1-8. Clone 5 has an incorrect amplicon size and should not be sent for sequencing.

Figure 5. Representative sequencing results for Snrpn from a single MII oocyte. Snrpn is methylated in oocytes. Black circles indicate methylated CpGs. White circles indicate unmethylated CpGs. CpG number and placement is representative for a B6 strain female mouse. a) Expected sequencing results for Snrpn from a single MII oocyte. Only a single strand of DNA should amplify in all five clones. Oocytes with a single methylation pattern and identical non-CpG conversion pattern should be included in analyses (percent conversion of non-CpGs indicated to the right was calculated as the number of non-CpG cytosines converted to thymine as a percentage of total non-CpG cytosines). b) Sequencing results for Snrpn from a single MII oocyte with cumulus cell contamination. Note the dissimilarity between methylation states and conversion patterns indicating multiple strand amplification. c) Sequencing results for Snrpn from a single MII oocyte with both chromosome copies or polar body inclusion.
Access restricted. Please log in or start a trial to view this content.
Dyskusje
This single oocyte assay contains many steps with a number that are critical and require special care. The first is oocyte washing. It is particularly important to wash each oocyte multiple times in fresh medium drops following hyaluronidase treatment to remove as many cumulus cells as possible. Moreover, when transferring oocytes to acidic tyrode's solution for zona pellucida removal make sure surrounding medium is clear of cumulus cells. The oocyte is very sticky following zona removal, and any surrounding cumulus cell...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
The authors have nothing to disclose.
Podziękowania
This work was supported by the University of Western Ontario, the Department of Obstetrics and Gynaecology; and a grant ER06-02-188 from the Ministryof Research and Innovation, Early Researcher Award. MMD was supported by a CIHR Training Program in Reproduction, Early Development and the Impact on Health (REDIH) Graduate Scholarship.
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| Oocyte Collection | |||
| Hyaluronidase | Sigma | H4272 | |
| Acidic Tyrode | Sigma | T1788 | |
| Proteinase K | Sigma | P5568 | |
| 10% IGEPAL | Bioshop | NON999.500 | |
| Lysis Solution | |||
| Tris pH 7.5 | Bioshop | TRS001.5 | |
| LiCl | Sigma | L9650 | |
| EDTA pH 8.0 | Sigma | E5134 | |
| LiDS | Bioshop | LDS701.10 | |
| DTT | Invitrogen | P2325 | |
| SDS Lysis Buffer | |||
| TE pH 7.5 | Bioshop(Tris)Sigma (EDTA) | TRS001.5 E5134 | |
| 10% SDS | Bioshop | SDS001.500 | |
| Bisulfite Conversion | |||
| Sodium Hydroxide | Sigma | S8045 | |
| Sodium Hydrogensulfite (Sodium Bisulfite) | Sigma | 243973 | |
| Hydroquinone | Sigma | H9003 | |
| Low Melting Point (LMP) Agarose | Sigma | A9414 | |
| Mineral Oil | Sigma | M8410 | |
| M2 Medium | Sigma | M7167 | |
| GIBCO Distilled water | Invitrogen | 15230-196 | |
| Autoclaved double distilled (dd) water | |||
| PCR | |||
| Illustra Hot Start Mix RTG | GE Healthcare | 28-9006-54 | |
| 240 ng/ml yeast tRNA | Invitrogen | 15401-011 | |
| Inner and outer nested primers | Sigma | ||
| Ligation | |||
| Promega pGEM-T Easy Vector | Fisher Scientific | A1360 | |
| TA Cloning | |||
| Competent E.coli cells | Zymo Research Corp. | T3009 | |
| Equipment | |||
| Dissecting Microscope | |||
| 70 °C and 90 °C Heat Blocks | |||
| 37 °C and 50 °C Waterbaths (42 °C for transformations) | |||
| Rocker | |||
| PCR machine |
Odniesienia
- Jaenisch, R., Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245-254 (2003).
- Rodenhiser, D., Mann, M. Epigenetics and human disease: translating basic biology into clinical applications. CMAJ. 174, 341-348 (2006).
- Frommer, M. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 89, 1827-1831 (1992).
- Clark, S. J., Harrison, J., Paul, C. L., Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990-2997 (1994).
- Feil, R., Charlton, J., Bird, A. P., Walter, J., Reik, W. Methylation analysis on individual chromosomes: improved protocol for bisulphite genomic sequencing. Nucleic Acids Res. 22, 695-696 (1994).
- Raizis, A. M., Schmitt, F., Jost, J. P. A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Anal. Biochem. 226, 161-166 (1995).
- Patterson, K., Molloy, L., Qu, W., Clark, S. DNA Methylation: Bisulphite Modification and Analysis. J. Vis. Exp. (56), e3170(2011).
- Olek, A., Oswald, J., Walter, J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24, 5064-5066 (1996).
- Mann, M. R. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 131, 3727-3735 (2004).
- Market-Velker, B. A., Zhang, L., Magri, L. S., Bonvissuto, A. C., Mann, M. R. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum. Mol. Genet. 19, 36-51 (2010).
- Meng, L. H., Xiao, S. Q., Huang, X. F., Zhou, Y., Xu, B. S. A study on bisulfite sequencing method for methylation status of imprinted genes in single human oocytes. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 25, 289-292 (2008).
- Denomme, M. M., Zhang, L., Mann, M. R. Embryonic imprinting perturbations do not originate from superovulation-induced defects in DNA methylation acquisition. Fertil. Steril. 96, 734-738 (2011).
- Tahiliani, M. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 324, 930-935 (2009).
- Hajj, N. E. l Limiting dilution bisulfite (pyro)sequencing reveals parent-specific methylation patterns in single early mouse embryos and bovine oocytes. Epigenetics. 6, 1176-1188 (2011).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone