A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Selecting and Isolating Colonies of Human Induced Pluripotent Stem Cells Reprogrammed from Adult Fibroblasts
In This Article
Summary
We present a protocol for efficient reprogramming of human somatic cells into human induced pluripotent stem cells (hiPSC) using retroviral vectors encoding Oct3/4, Sox2, Klf4 and c-myc (OSKM) and identification of correctly reprogrammed hiPSC by live staining with Tra-1-81 antibody.
Abstract
Herein we present a protocol of reprogramming human adult fibroblasts into human induced pluripotent stem cells (hiPSC) using retroviral vectors encoding Oct3/4, Sox2, Klf4 and c-myc (OSKM) in the presence of sodium butyrate 1-3. We used this method to reprogram late passage (>p10) human adult fibroblasts derived from Friedreich's ataxia patient (GM03665, Coriell Repository). The reprogramming approach includes highly efficient transduction protocol using repetitive centrifugation of fibroblasts in the presence of virus-containing media. The reprogrammed hiPSC colonies were identified using live immunostaining for Tra-1-81, a surface marker of pluripotent cells, separated from non-reprogrammed fibroblasts and manually passaged 4,5. These hiPSC were then transferred to Matrigel plates and grown in feeder-free conditions, directly from the reprogramming plate. Starting from the first passage, hiPSC colonies demonstrate characteristic hES-like morphology. Using this protocol more than 70% of selected colonies can be successfully expanded and established into cell lines. The established hiPSC lines displayed characteristic pluripotency markers including surface markers TRA-1-60 and SSEA-4, as well as nuclear markers Oct3/4, Sox2 and Nanog. The protocol presented here has been established and tested using adult fibroblasts obtained from Friedreich's ataxia patients and control individuals 6, human newborn fibroblasts, as well as human keratinocytes.
Protocol
1. Virus production and transduction
- Plate Phoenix Ampho cells at a density of ~7-8x106 per 10 cm plate in 10 ml of DMEM medium (DMEM high glucose, 10% FBS heat inactivated, 2 mM L-glutamine, no antibiotics). Place in the incubator and culture overnight at 37°C, 5% CO2.
- The next day transfect Phoenix cells using 12 μg of a vector encoding either Oct3/4, Sox2, Klf4, c-myc, or GFP gene (Addgene plasmids 17217, 17218, 17219, 17220) and 35 μl Fugene 6. Prepare transfection mix in 500 μl of DMEM medium (DMEM high glucose, 2 mM L-glutamine, no FBS and antibiotics). Incubate 20 min. Gently pipette DNA complexes into the 10 cm plates containing 70-80% confluent Phoenix cells.
- Replace media 6 - 8 h post-transfection with DMEM medium (DMEM high glucose, 10% FBS heat inactivated, 2 mM L-glutamine) containing penicillin and streptomycin.
- Subsequently, collect virus-containing media 4 times in 12 h intervals, combine all portions and filtered using a 0.45 μm filter to remove detached cells and debris (media containing viral particles can be kept in the refrigerator for 2 weeks without losing infectious activity, however freezing is not recommended).
- For retroviral transduction, plate human adult fibroblasts (passage 10, Coriell Laboratories) from a frozen stock 24 h prior to the last day of retroviral media collection. Seed the cells on 6-well gelatin covered plates at the density 1x105 cells per well in DMEM high glucose, 10% FBS, 2 mM L-glutamine, penicillin, streptomycin and non-essential amino acids.
- The next day substitute DMEM media with media containing viral particles obtained from Phoenix cells. Add viral media (1 ml of each Oct4, Sox2, Klf4, and c-myc media, 4 ml total) supplemented with 6 μg/ml of polybrene into each well of the 6-well plate of the fibroblast cultures and centrifuge at 1600g for 1h at 20°C. Approximately 12 h after the transduction, replace the viral media with fibroblast culture medium. Perform viral infection and centrifugation 3 times in 24 h intervals.
- Culture fibroblasts in DMEM media for 48 h after last infection. Check the efficiency of the infection by parallel transductions with a retroviral media expressing GFP. Figure 1 illustrates the efficiency of centrifugation-facilitated retroviral infection of human fibroblast.
2. Reprogramming
- Prepare 6 well plates with γ-irradiated MEF feeder layers by seeding MEF cells at a density of ~2x105 cells per well of the gelatin treated 6 well plate in fibroblast growth medium. The following day split infected human fibroblasts using 0.05% trypsin/EDTA and seed them at the density of ~1.2x104 per well in the fibroblasts growth medium.
- The next day replace media with hES media (DMEM/F12, 20% Knockout Serum Replacement, non-essential amino acids, penicillin/streptomycin, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 20 ng/ml basal Fibroblast Growth Factor (bFGF), supplemented with 0.5 mM sodium butyrate for the initial 7 days of the reprogramming. Change media daily.
- Monitor morphological changes in transduced cells daily. Colonies of hiPS-like cells will start to emerge approximately 5 - 10 days after transferring the transduced fibroblasts onto feeder cells. The hiPSC colonies are ready to be isolated approximately 14 - 28 days after plating them on the MEF feeder cells.
3. Isolation of hiPSC colonies
- A day before picking the hiPSC colonies prepare 24-well plates with γ-irradiated MEF feeder cells (~4x104 cells per well) in fibroblasts growth medium. At this stage hiPSC colonies can also be transferred to feeder free culture using Matrigel and mTeSR1 medium.
- Prior to isolation of hiPSC colonies prepare MEF plates by removing fibroblasts growth medium and rinsing MEFs with PBS to remove traces of FBS. Subsequently add 0.5 ml hES medium (or mTeSR1 medium in the case of Matrigel coated wells) with 10 μM ROCK inhibitor Y27632 to each well 7,8.
- If necessary, remove the fibroblasts surrounding the hiPSC colonies with a 21 gauge needle under a microscope mounted in a laminar flow hood (Figure 2A - C). Rinse the plates with PBS and add fresh hES media containing the Tra-1-81 StainAlive specific antibody (1:200, Stemgent). After 30 min., replace media containing the antibody with fresh hES media supplemented with 10 μM ROCK inhibitor. Examine the plates under the fluorescent microscope and mark Tra-1-81 positive colonies using an objective marker (Figure 2D).
- Cut the Tra-1-81 positive hiPSC colonies under a microscope in a laminar hood, into several small pieces using a 21 gauge needle as shown in Figure 2E and F.
- Using an automatic pipette (P200) transfer fragments of hiPSC colonies into individual wells of the 24-well plate with MEFs or Matrigel. Avoid transferring non-hiPS cells. Place the plates in a 37°C, 5% CO2 incubator and allow the hiPSC colonies to attach for 24-36 h.
- Change hES or mTeSR1 (for colonies grown on Matrigel) media daily. The hiPSC colonies with correct hES-like morphology will be visible 48 h after the initial transfer onto a 24-well plate.
- Manually passage hiPSC colonies every 6 - 8 days onto a 12-well and subsequently on a 6-well plate.
- After clonal expansion and establishing the hiPSC lines, analyze expression of pluripotency markers TRA-1-60, SSEA-4, Oct3/4, Sox2 and Nanog using immunocytochemistry as shown in Figure 3. Evaluate genomic integrity and differentiation potential of the obtained lines using karyotype and teratoma formation analyses 6,9.
4. Representative Results
Efficient transduction with retrovirus-containing media is critical for successful reprogramming. It is recommended to conduct the entire transfection/infection procedure using a GFP expressing virus every single reprogramming experiment to monitor the efficiency as shown in Figure 1. The titer of the GFP expressing virus determined, as described in 10, through the transduction of human fibroblasts using non-concentrated viral media was typically in the range of 0.5 - 5 x 107 viral particles per ml (vp/ml).
Fibroblasts change morphology as early as 2 days after the last infection. Trypsinized human fibroblasts should be carefully counted prior to seeding them on the MEF feeder cells. It is recommended to seed cells at 3 different densities (6x103, 1.2x104, 2.5x104 per single well of a 6 well plate) since each cell line demonstrates different growth characteristic and seeding density is critical for the efficiency of reprogramming. Sodium butyrate used for the initial 7 - 14 days of reprogramming increases the efficiency of hiPSC formation approximately 5 fold. Frequently, especially when infected fibroblasts were seeded at higher density, the fibroblast-like cells can overgrow a culture dish and cover hiPSC colonies as shown in Figure 2A. In this case, the fibroblast layer can be carefully lifted and removed to uncover hiPSC colonies (Figure 2B). Subsequently, hiPSC colonies should be rinsed with hES media and stained with Tra-1-81 antibody. Depending on the fibroblast cells, approximately 20 - 40% of colonies demonstrating with iPSC-like morphology do not stain with Tra-1-81 antibody. Identified hiPSC colonies can be transferred from a plate within next 12 - 24h. Prolonged incubation will result in rapid differentiation of hiPSCs. Colonies can be manually passaged onto either MEF feeder cells or Matrigel-coated plates. After expansion established hiPSC clones should be tested using immunocytochemistry (ICC) for the expression of pluripotency markers as demonstrated in Figure 3. Additionally, a detailed molecular characterization of the generated iPS cell lines should include: analyses of the pluripotency gene expression using RT-PCR, demonstration of the DNA demethylation at the promoters of pluripotency genes and analyses of the transgenes silencing 9.
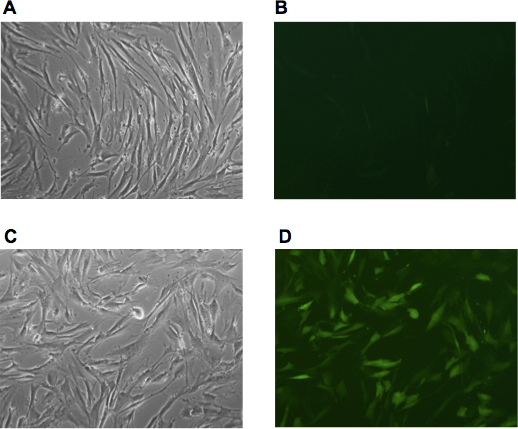
Figure 1. The efficiency of viral transduction determined using GFP expressing retrovirus. (A, B) Human adult fibroblasts derived from Friedreich's ataxia patient (GM03665, Coriell Repository) were visualized after two consecutive infections with GFP retroviral media. (C, D) Human fibroblasts were infected with the same batch of the GFP retroviral media followed by centrifugation of the cells directly on the 6-well plates for 1h at 1600g. Images were captured 48h after infection.
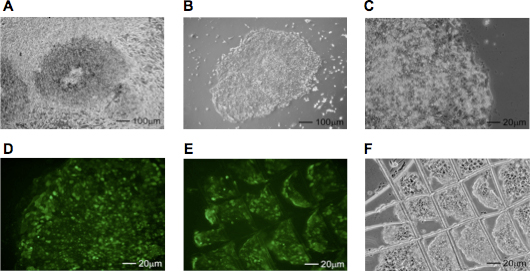
Figure 2. Identification and isolation of hiPSC colonies. (A) Phase contrast image of a plate containing hiPSCs surrounded by fibroblasts. The cells were cultured for 21 days on hES media. (B, C) The same hiPSC colony after removal of the surrounding fibroblast layer. (D) Correctly reprogrammed hiPSC colonies are identified by live staining with Tra-1-81 surface marker antibody, cut using a sterile needle (E, F) and transferred to separate wells of a 24-well plate.
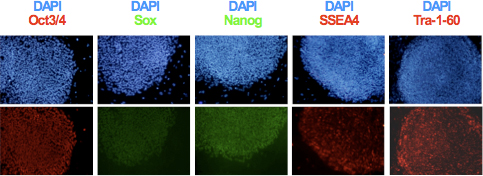
Figure 3. Expression of the pluripotency specific markers Oct3/4, Nanog, Sox2, SSEA4 and Tra-1-60 in hiPSCs was determined by immunocytochemistry.
Discussion
Studying human diseases, especially neurological and neurodegenerative, has been particularly challenging due to the inaccessibility of adequate human cellular models. The ability to reprogram easily obtainable somatic cells into induced pluripotent stem cells and the potential to differentiate them into diverse cell types opened a possibility to create cellular models of genetic diseases. In addition, iPSCs hold a great promise in the future of regenerative medicine. Therefore, it is essential to develop and optimize re...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by Friedreich’s Ataxia Research Alliance and a pilot grant from Arnold Family Foundation and The Center for Stem Cells and Developmental Biology at M.D. Anderson Cancer Center.
Materials
| Name | Company | Catalog Number | Comments |
| DMEM | Invitrogen | 11965 | |
| DMEM/F12 | Invitrogen | 11330 | |
| KSR | Invitrogen | 10828 | |
| Non-essential aminoacids | Invitrogen | 11140 | |
| Sodium butyrate | Sigma-Aldrich | B5887 | |
| Y27632 | Stemgent | 04-0012 | |
| bFGF | Stemgent | 03-0002 | |
| Tra-1-81 antibody | Stemgent | 09-0069 | |
| Oct3/4 antibody | Santa Cruz Biotechnology, Inc. | sc-8628 | |
| Nanog antibody | Cell Signaling Technology | 4903S | |
| Tra-1-60 antibody | EMD Millipore | MAB4360 | |
| Sox2 antibody | Cell Signaling Technology | 3579S | |
| SSEA4 | EMD Millipore | MAB4304 | |
| CF1 MEFs | Globalstem | GSC-6201G | |
| Objective marker | Nikon Instruments | MBW10010 | |
| Matrigel | BD Biosciences | 354277 | |
| mTeSR1 | Stem Cell Technologies | 05850 | |
| β-mercapt–thanol | Sigma-Aldrich | M7522 | |
| Fugene 6 | Roche Group | 11814443001 | |
| polybrene | Sigma-Aldrich | H9268 | |
| Object marker | Nikon Instruments | MBW10010 |
References
- Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131, 861-872 (2007).
- Mali, P., Chou, B. K., Yen, J., Ye, Z., Zou, J., Dowey, S., Brodsky, R. A., Ohm, J. E., Yu, W., Baylin, S. B., Yusa, K., Bradley, A., Meyers, D. J., Mukherjee, C., Cole, P. A., Cheng, L. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 28, 713-720 (2011).
- Wernig, M., Meissner, A., Foreman, R., Brambrink, T., Ku, M., Hochedlinger, K., Bernstein, B. E., Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 448, 318-324 (2007).
- Lowry, W. E., Richter, L., Yachechko, R., Pyle, A. D., Tchieu, J., Sridharan, R., Clark, A. T., Plath, K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 105, 2883-2888 (2008).
- Dick, E., Matsa, E., Bispham, J., Reza, M., Guglieri, M., Staniforth, A., Watson, S., Kumari, R., Lochmuller, H., Young, L., Darling, D., Denning, C. Two new protocols to enhance the production and isolation of human induced pluripotent stem cell lines. Stem Cell Res. 6, 158-167 (2011).
- Ku, S., Soragni, E., Campau, E., Thomas, E. A., Altun, G., Laurent, L. C., Loring, J. F., Napierala, M., Gottesfeld, J. M. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell. 7, 631-637 (2010).
- Emre, N., Vidal, J. G., Elia, J., O'Connor, E. D., Paramban, R. I., Hefferan, M. P., Navarro, R., Goldberg, D. S., Varki, N. M., Marsala, M., Carson, C. T. The ROCK inhibitor Y-27632 improves recovery of human embryonic stem cells after fluorescence-activated cell sorting with multiple cell surface markers. PLoS One. 5, e12148-e12148 (2011).
- Watanabe, K., Ueno, M., Kamiya, D., Nishiyama, A., Matsumura, M., Wataya, T., Takahashi, J. B., Nishikawa, S., Muguruma, K., Sasai, Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681-686 (2007).
- Maherali, N., Hochedlinger, K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 3, 595-605 (2008).
- Yu, J., Thomson, J. A., Lanza, R. Induced Pluripotent Stem Cell Derivation. Essentials of Stem Cell Biology. , 331-337 (2009).
- Yu, J., Hu, K., Smuga-Otto, K., Tian, S., Stewart, R., Slukvin, I. I., Thomson, J. A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 324, 797-801 (2009).
- Woltjen, K., Michael, I. P., Mohseni, P., Desai, R., Mileikovsky, M., Hamalainen, R., Cowling, R., Wang, W., Liu, P., Gertsenstein, M., Kaji, K., Sung, H. K., Nagy, A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 458, 766-770 (2009).
- Zhou, W., Freed, C. R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 27, 2667-2674 (2009).
- Warren, L., Manos, P. D., Ahfeldt, T., Loh, Y. H., Li, H., Lau, F., Ebina, W., Mandal, P. K., Smith, Z. D., Meissner, A., Daley, G. Q., Brack, A. S., Collins, J. J., Cowan, C., Schlaeger, T. M., Rossi, D. J. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 7, 618-630 (2010).
- Kim, D., Kim, C. H., Moon, J. I., Chung, Y. G., Chang, M. Y., Han, B. S., Ko, S., Yang, E., Cha, K. Y., Lanza, R., Kim, K. S. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 4, 472-476 (2009).
- Han, S. S., Williams, L. A., Eggan, K. C. Constructing and deconstructing stem cell models of neurological disease. Neuron. 70, 626-644 (2011).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved