A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
RNAi Screening to Identify Postembryonic Phenotypes in C. elegans
In This Article
Summary
We describe a sensitized method to identify postembryonic regulators of protein expression and localization in C. elegans using an RNAi-based genomic screen and an integrated transgene that expresses a functional, fluorescently tagged protein.
Abstract
C. elegans has proven to be a valuable model system for the discovery and functional characterization of many genes and gene pathways1. More sophisticated tools and resources for studies in this system are facilitating continued discovery of genes with more subtle phenotypes or roles.
Here we present a generalized protocol we adapted for identifying C. elegans genes with postembryonic phenotypes of interest using RNAi2. This procedure is easily modified to assay the phenotype of choice, whether by light or fluorescence optics on a dissecting or compound microscope. This screening protocol capitalizes on the physical assets of the organism and molecular tools the C. elegans research community has produced. As an example, we demonstrate the use of an integrated transgene that expresses a fluorescent product in an RNAi screen to identify genes required for the normal localization of this product in late stage larvae and adults. First, we used a commercially available genomic RNAi library with full-length cDNA inserts. This library facilitates the rapid identification of multiple candidates by RNAi reduction of the candidate gene product. Second, we generated an integrated transgene that expresses our fluorecently tagged protein of interest in an RNAi-sensitive background. Third, by exposing hatched animals to RNAi, this screen permits identification of gene products that have a vital embryonic role that would otherwise mask a post-embryonic role in regulating the protein of interest. Lastly, this screen uses a compound microscope equipped for single cell resolution.
Protocol
1. Screening strain construction
The careful design of the screening strain is critical for the success of the screen and has been described elsewhere3. For some researchers, using a strain that expresses a visible product from a transgene is needed for the experiment. Many strains harboring integrated transgenes are available from the CGC or individual researchers. If a transgenic strain is required for the screen but is not available, then it can be generated using a published method like bombardment4, UV/TMP5, or Mos transposon insertion6. In order to visualize our protein of interest, we inserted the gfp-coding sequence in frame with the mature cDNA sequence (we used dbl-1 sequence). Because this GFP fusion protein is not visible by dissecting scope, we used a coinjection marker that was visible and did not affect the viewing of our protein of interest (ttx-3p::rfp, for a review of several other standard markers, see 7). We then created an integrated transgene from the extrachromosomal array. We found that bombardment of the transgene yielded a low-copy number integrated line that neither rescued the dissecting microscope phenotype nor produced visible levels of GFP-tagged product (Beifuss and Gumienny, unpublished). UV/TMP integration of a multiple copy extrachromosomal array originally made by injection8,9 yielded visible, rescuing levels of transgene product (Figure 3A). Western blot with anti-GFP antibody confirmed that the transgene (allele name texIs100) is expressed and processed correctly (Beifuss and Gumienny, unpublished). Integrated transgenes should be outcrossed five times to remove extraneous background mutations regardless of source.
For our screen, we wanted the only protein of interest to be the transgenic, tagged form. Therefore, we removed the functional endogenous gene by introducing a loss-of-function mutant dbl-1 allele.
Lastly, we sensitized the strain to the effects of RNAi. The RNAi from the library will, for many genes, produce a more severe reduction in gene product if animals contain a mutation that sensitizes the animals to the effects of RNAi10,11 The tissue(s) of interest should be considered when choosing an appropriate RNAi sensitizing background11. We used the canonical rrf-3(pk1426) allele to make our screening strain. RRF-3 is an RNA-directed RNA polymerase (RdRP) homolog that normally inhibits somatic RNAi10. Mutations in other RNAi hypersensitizing genes like eri-1 or eri-1 lin-15b can be used instead to increase the effectiveness of the RNAi11-13.
The screening strain we made has the genotype rrf-3(pk1426); texIs100; dbl-1(nk3).
2. Choosing and preparing an RNAi library
Commercially available C. elegans cDNA libraries represent about 55% or 87% of the predicted genes in C. elegans individually (Vidal lab or Ahringer lab libraries, respectively). Individual clones are available for purchase (Open Biosystems, Geneservice, Ltd.). We chose the ORF-RNAi library constructed by the Vidal lab (Open Biosystems) because its clones are mostly full-length cDNAs Gateway cloned and ready for downstream applications (Invitrogen Corporation, Carlsbad, CA). The original Ahringer lab genomic cDNA library contains cDNA fragments that are not as widely useful for downstream characterization experiments14,15. Both libraries use a vector that contains two T7 promoters flanking the insert (Figure 1). The constructs are grown in bacterial strain HT115, which expresses T7 polymerase upon induction by isopropyl-β-D-thiogalactopyranoside (IPTG). Bacteria containing library clones are induced with IPTG to produce dsRNA (see Step 3.4).
Duplicate the entire library upon receipt and use the duplicate for all experiments. The original and duplicate libraries should be stored in different -80°C freezers that are connected to separate electrical lines.
3. Preparation of bacteria with library clones
- (Within two months of the experiment) Streak bacteria from selected feeding library clones, as well as positive and negative controls, on labeled LB- carbenicillin/ tetracycline plates (we grow 8 cultures per 100 mm plate) and incubate at 37°C overnight (>16 hours) (Figure 1). For each 24-well plate, we use two controls. The positive control, bli-4 (K04F10.4), produces a dose-dependent post-embryonic pheontype16,17. The negative control, C06C3.5, is a predicted pseudogene (WormBase.org) that causes no observed phenotypes (Beifuss and Gumienny, unpublished).
- (Within two months of the experiment, preferably sooner) Prepare 24-well Nematode Growth Medium (NGM) plates containing 25 – 50 μg/ml carbenicillin and 1mM IPTG. Store at 4°C.
- (Day 1) For each clone, inoculate 2 ml LB media containing 50 μg/ml carbenicillin with a single colony from the streaked plate(s) (in Step 3.1) using standard sterile technique. Incubate at 37°C overnight (>16 hours) in a shaker set to 220 – 240 rpm (Figure 1). The solution should be cloudy after 16 hours.
- (Day 2) The next morning, induce double stranded RNA (dsRNA) production by adding IPTG to the cultures to a final concentration of 1mM. Incubate cultures at 37°C, 220 – 240 rpm, for an additional 4 – 5 hours. This step ensures a more consistent and robust RNAi-induced knockdown of gene product than relying solely on the IPTG in the agar of the NGM plate (in Step 3.5).
- Spot 30 μl of each induced bacterial culture into separate wells of a 24-well NGM RNAi plate containing 25 – 50 μg/ml carbenicillin and 1mM IPTG (Figure 1, see Figure 4 for template used to track RNAi experiments). Put plates, uncovered, in a sterile flow hood for 20 minutes or until the bacterial spots are dry. Do not overdry. Overdrying causes the edges of the agar to pull away from the plate and animals will be irretrievable if they crawl into the resulting crevices.
4. Preparation of nematodes
Start with a staged population of nematodes. Staging animals decreases the chance that differences observed between the control and the experimental RNAi trials are simply due to differences in developmental stage of the animals (however, if the RNAi causes a developmental delay, this should be noted by the screener (Figure 2)). Furthermore, starting the RNAi experiment with L1 larvae obviates potential confounding effects of embryonic lethality by RNAi of genes that may also play postembryonic roles of interest.
Staging is accomplished by first bleaching a mixed-stage population of the screening strain, which only eggshell-protected embryos survive. This stages animals to within about 12 hours at 20°C or about 18 hours at 16°C18. To more tightly stage animals, arrest animals in the first larval stage (L1) by letting embryos hatch in M9 medium without food. Starved L1 animals placed on food resume growth from the same starting age19.
- (Day 1) Use three 100 mm plates of fed, clean screening strain animals that contain gravid adults and embryos. Wash animals from plates using sterile M9 by pipeting liquid across the plate surface gently to loosen animals and embryos. Transfer the wash into a sterile 15 ml tube with screw cap.
- If the wash is greater than 3.5 ml, spin down the animals (~3000 rpm for 30 seconds) and remove liquid to 3.5 ml. If the wash is less than 3.5 ml, add H20 or M9 to the tube to total 3.5 ml.
- Wear appropriate protective gear (lab coat, gloves, and goggles) while handling chemicals. Mix 0.5 ml 5 N NaOH with 1 ml fresh bleach (5% sodium hypochlorite, less than 1 year old). Add this mixture to the 3.5 ml nematode wash. Start a timer counting up. This process should not take more than 10 minutes. If it does, then embryos will be endangered and yield may be reduced, or the bleach is old and should be replaced.
- Vortex the tube for about 10 seconds. Repeat the vortexing every two minutes for 4 to 10 minutes. After each vortexing, observe the solution under a dissecting scope for animal carcasses.
- When embryos are released and adults are fragmented (6 – 8 minutes), pellet the embryos by spinning the tube in a table top centrifuge (~3000 rpm, 30 seconds).
- Aspirate as much of the supernatant as possible without disturbing the pellet. Be careful, not greedy.
- Add sterile M9 to ~ 10ml. Vortex briefly or shake well to resuspend the pellet.
- Spin and remove supernatant again. If the smell of bleach can be detected in the tube, repeat rinse as necessary.
- Resuspend embryos in 10 – 15 ml sterile M9 and transfer to a small (25 ml) Erlenmeyer flask (for aeration). Shake at the proper temperature for the strain and experiment (15 – 25°C) overnight (> 16 hours at 20°C). In this time, the animals will all hatch and arrest in the L1 diapause.
- (Day 2) Spin down L1 stage animals in a 15 ml tube and resuspend in 0.5 – 1 ml M9 (~3000 rpm, 30 sec.). Place 5 μL solution on a separate plate and count the number of animals. Make adjustments to the volume, if needed, to yield 30 worms/ 3 – 10 μL solution.
- Spot approximately 30 hypochlorite synchronized L1 stage animals onto the bacteria in each well of the prepared 24-well plate. Too many more than 30 animals per well may result in the population starving before reaching maturity. Let the plate dry with the lids askew. Replace the lid and secure it with a rubber band. Invert the plate and place it at the appropriate temperature for the time needed to score at the desired stage. Because rrf-3(pk1426) is a temperature sensitive allele, our screening strain animals were grown at 15 – 17°C for three to four days to observe L4 to adult stage animals (Figure 1).
5. Observation of nematodes
(Day 5) Once the nematodes have grown to the desired stage (Figure 2A), confirm that the positive and negative controls produce the expected phenotypes by using a dissecting microscope. We use bli-4 as a control for RNAi efficacy, because it produces dose-dependent post-embryonic defects that range from blistered adults (mild RNAi phenotype, not shown) to developmentally arrested, tiny larvae (strong, expected RNAi phenotype) (Figure 2B). Then observe the animals in each experimental well using a dissecting microscope and note obvious abnormal phenotypes induced by RNAi (Figure 2C). After the controls are confirmed and gross phenotypes noted, screen the RNAi experiments for phenotypes of interest. We use a compound microscope equipped with fluorescence and a 63x objective to observe at least five animals from each RNAi experiment, beginning with controls (Figure 3).
- Prepare a slide (standard 75 x 25 mm) by making a 4% agar pad (4% agar in H2O). To ensure a uniform thickness of agar pad, place a clean glass slide between two spacers (each spacer is made of a microscope slide with two layers of lab tape on it). Pipette about 100 μL (two to three drops from a glass Pasteur pipet) of molten 4% agar onto the center of the clean glass slide. Form an agar pad by covering the molten agar using an additional slide placed quickly but gently on top of the spacers and the molten agar. Expose the agar pad by gently sliding the glass slides apart, leaving the pad adhering to one slide.
- Place a ~5 μL drop of anesthetic (levamisole or sodium azide, for example) on the agar pad. Mount 8 – 12 animals in the anesthetic. Cover with a 22 x 22 mm #1.5 coverslip (or the thickness of coverslip best suited for the compound microscope) and observe animals using a compound microscope.
- Record gene, number of animals observed, and phenotype results for each RNAi experiment. For a screen, make and use a standard template (see example, Figure 5).
- Verify the RNAi-produced phenotype by another, secondary phenotype, if possible, and by repeating the experiment. (For example, the example screen shown used alteration of GFP-tagged DBL-1 localization as a primary screen and body size as a secondary screen20.) Bacteria can be used from the same streak (stored at 4°C) for up to two months. Older streaks may result in poor growth in liquid overnight cultures and should be restreaked first.
- Sequence at least one end of the cDNA insert to confirm that the insert matches the library label (the Ahringer library has had a reliability analysis performed; see http://biocompute.bmi.ac.cn/CelRNAi/)21. Recommended primers (for inserts from either library) anneal on sites in the L4440-derived vector and flank the insertion site: 5'-AGCGAGTCAGTGAGCGAG-3' and 5'-GTAAAACGACGGCCAGT-3' (M13f20 primer).
Using this method, a single person can reasonably stage and observe two to four sets of experiments every week. For instance, animals staged on Monday and Tuesday can be observed Thursday and Friday, respectively, and can again be staged on Friday and Saturday for Monday and Tuesday observation. Depending on the phenotype(s) screened each day, a set may comprise one 24-well plate by compound microscopy or more if the phenotype is quickly identified. Thus, at least 88 different RNAi experiments can be observed in one week, taking into account positive and negative controls and rate of screening. A dissecting scope screen could be performed much faster, without the need for mounting animals on slides for viewing. Multiple people could also increase a lab's throughput by staggering schedules and/or using multiple microscopes. An alternative method for growing animals on a thin film of agar and transferring a slice of the agar containing animals directly to a slide for viewing may save time. This variation was successfully used for screening male tail abnormalities at 400x magnification22.
6. Representative Results
Examples of normal and altered localization of GFP-tagged DBL-1 are shown in Figure 3. Normal expression of GFP-tagged DBL-1 includes ventral nerve cord cell bodies and a row of punctae (Figure 3A). DBL-1 is severely attenuated when animals are fed RNA that prevents dbl-1 mRNA translation (Figure 3B). dbl-1(RNAi) also produces small animals, the secondary screen in this example (data not shown). Genes that affect DBL-1 localization are readily identified by RNAi using a strain designed for this screen (Figure 3C).

Figure 1. Screen scheme to identify extracellular regulators of DBL-1 signaling. Bacteria from the feeding library are grown overnight, induced with IPTG, and incubated at 37°C for an additional 4 hours to allow the bacteria to produce double stranded RNA (dsRNA). 30 μl of the induced bacterial culture is spotted per well onto a 24-well NGM (nematode growth medium) plate containing 25 μg/ml carbenicillin and 1mM IPTG and allowed to dry in a sterile flow hood. We stage first larval stage (L1) larvae by letting hypochlorite-treated embryos hatch in media without food, which induces an L1 diapause (arrested development). Approximately 30 synchronized L1 stage animals are plated onto each well containing bacteria from the RNAi library, which allows staged animals to resume growth and to consume the dsRNA created by the bacteria23. After 72 hr at 15°C, the young adult worms are screened for a visible phenotype. At this age, body size defects are evident and fluorescence is bright from texIs100 transgene expression.
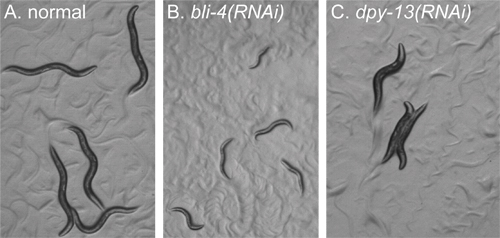
Figure 2. Examples of phenotypes to identify before screening. All images of animals in a plate well were taken at the same magnification using a dissecting microscope. Animals were all imaged about 72 hours after plating as starved L1 larvae. All images were treated identically. A) RNAi of a gene that gives no gross morphological defects. Animals appear wild type. B) RNAi of bli-4. Animals display arrested development, and are tiny compared to the animals in panel A. C) RNAi of dpy-13. Animals are at the same stage of development as animals in panel A, but display a "dumpy" body morphology.
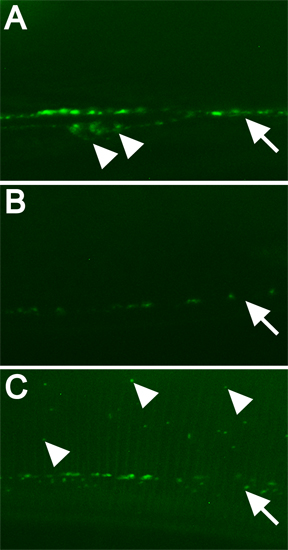
Figure 3. Examples of fluorescently tagged protein and how RNAi of specific genes alters localization pattern. All images were taken at 630x magnification with spinning disk confocal microscopy, 5 sec. exposure. Scale bar = 10 μm. All images were treated identically. Open arrowheads indicate cell bodies. Arrows mark line of punctae. Filled arrowheads indicate some aberrantly localized GFP-tagged DBL-1. A) RNAi-fed pseudogene C06C3.5 ("wild-type"). B) dbl-1(RNAi) control. C) RNAi of a gene required for normal localization of GFP-tagged DBL-1.
Figure 4. Template example for tracking RNAi experiments (library clones) in 24-well plates. This template can be used to create a permanent record of the experiments, unlike directly labeling plates.
Figure 5. Template example for recording RNAi phenotypes. Expand as needed.
Discussion
The RNAi screening method presented here enables a sensitive and rapid analysis of gene products required for a normal (or transgenic) postembryonic phenotype. The example shown is a screen for genes involved in the subcellular localization of a fluorescently tagged protein. However, this protocol can be modified to identify genes affecting other postembryonic phenotypes of interest.
This method takes advantage of a candidate gene approach by using an RNAi library. Forward genetic screens usin...
Disclosures
We have nothing to disclose.
Acknowledgements
The authors would like to thank Dr. Rick Padgett (Waksman Institute, Rutgers University, NJ) for the gift of the dbl-1 cDNA and Dr. Christopher Rongo (Waksman Institute, Rutgers University, NJ) for an injection marker. Dr. Barth Grant's lab performed the gene gun bombardment for low copy number integration of the gfp-tagged dbl-1 construct. The René Garcia laboratory provided technical assistance during the creation of texIs100. The René Garcia, Robyn Lints, and Hongmin Qin laboratories provided productive advice. This work was funded by start-up funds from the TAMHSC Department of Molecular and Cellular Medicine. The compound scope and spinning disk confocal were purchased with funds provided by the department and the TAMHSC College of Medicine Office of the Dean.
Materials
| Name | Company | Catalog Number | Comments |
| NGM Agar | Nematode growth medium | IPM Scientific, Inc | Can be prepared following NGM agar protocol25 |
| M9 Medium | 22mM KH2PO4, 42mM Na2HPO4, 86mM NaCl, 1 mM MgSO4 | ||
| Agar-Agar | EMD Millipore | 1.01614.1000 | 2% in water for NGM plates. 4% in water for microscope slide pads (autoclave initially and microwave to melt thereafter). |
| Bacto Peptone | BD Biosciences | 211677 | 0.25% |
| IPTG | Research Products International Corp. | I56000-5.0 | 1 mM final concentration |
| carbenicillin | Research Products International Corp. | C46000-5.0 | 50 μg/ml working dilution |
| LB Broth Lennox | BD Biosciences | 240230 | 20 g/liter |
| tetracycline | Sigma-Aldrich | 268054 | 12.5 μg/ml working dilution |
| sodium hypochlorite | Any Supplier | 5% household bleach | Use fresh bleach. |
| sodium hydroxide | Any Supplier | CAS 1310-73-2 | 5 N stock |
| M9 medium | Wormlab Recipe Book | http://130.15.90.245/wormlab_recipe_book.htm#Commonlab | 26 |
| levamisol | Sigma-Aldrich | 31742 | 100 μM - 1 mM working dilution |
| sodium azide | Fisher Scientific | S227 | 10 mM in M9 working dilution |
| 24-well plate | Greiner Bio-One | 662160 | VWR distributor |
| microscope slides | Any Supplier | 75 x 25 x 1 mm | |
| microscope cover slips | Any Supplier | 22 x 22 mm No.1.5 | Use the thickness recommended by the microscope manufacturer. |
| compound microscope | Carl Zeiss, Inc. | A1m | Use objectives and filters to match the needs of the experiment. |
| media pump | Manostat Varistaltic pump | Kate model #72-620-000 | Use tubing and settings appropriate for the machine |
References
- Giacomotto, J., Segalat, L. High-throughput screening and small animal models, where are we. Br. J. Pharmacol. 160, 204-216 (2010).
- Lamitina, T. Functional genomic approaches in C. elegans. Methods in Molecular Biology. 351, 127-138 (2006).
- Boutros, M., Ahringer, J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 9, 554-566 (2008).
- Praitis, V., Casey, E., Collar, D., Austin, J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157, 1217-1226 (2001).
- Yandell, M. D., Edgar, L. G., Wood, W. B. Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 91, 1381-1385 (1994).
- Frokjaer-Jensen, C. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature Genetics. 40, 1375-1383 (2008).
- Giordano-Santini, R., Dupuy, D. Selectable genetic markers for nematode transgenesis. Cell. Mol. Life. Sci. , (2011).
- Berkowitz, L. A., Knight, A. L., Caldwell, G. A., Caldwell, K. A. Generation of Stable Transgenic C. elegans Using Microinjection. J. Vis. Exp. (18), e833 (2008).
- Mello, C., Fire, A. DNA transformation. Methods in Cell Biology. 48, 451-482 (1995).
- Simmer, F. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12, 1317-1319 (2002).
- Zhuang, J. J., Hunter, C. P. Tissue-specificity of Caenorhabditis elegans enhanced RNAi mutants. Genetics. , (2011).
- Samuelson, A. V., Klimczak, R. R., Thompson, D. B., Carr, C. E., Ruvkun, G. Identification of Caenorhabditis elegans genes regulating longevity using enhanced RNAi-sensitive strains. Cold Spring Harbor Symposia on Quantitative Biology. 72, 489-497 (2007).
- Wang, D. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 436, 593-597 (2005).
- Fraser, A. G. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 408, 325-330 (2000).
- Kamath, R. S. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421, 231-237 (2003).
- Peters, K., McDowall, J., Rose, A. M. Mutations in the bli-4 (I) locus of Caenorhabditis elegans disrupt both adult cuticle and early larval development. Genetics. , 129-195 (1991).
- Thacker, C., Peters, K., Srayko, M., Rose, A. M. The bli-4 locus of Caenorhabditis elegans encodes structurally distinct kex2/subtilisin-like endoproteases essential for early development and adult morphology. Genes & Development. 9, 956-971 (1995).
- Byerly, L., Cassada, R. C., Russell, R. L. The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev. Biol. 51, 23-33 (1976).
- Stiernagle, T. Maintenance of C. elegans. WormBook. , 1-11 (2006).
- Savage-Dunn, C. Genetic screen for small body size mutants in C. elegans reveals many TGFbeta pathway components. Genesis. 35, 239-247 (2003).
- Qu, W. Reliability analysis of the Ahringer Caenorhabditis elegans RNAi feeding library: a guide for genome-wide screens. BMC Genomics. 12, 1471-2164 (2011).
- Nelson, M. D. A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in Caenorhabditis elegans. PLoS Genetics. 7, e1002010 (2011).
- Timmons, L., Fire, A. Specific interference by ingested dsRNA. Nature. 395, 854 (1998).
- Sarin, S., Prabhu, S., O'Meara, M. M., Pe'er, I., Hobert, O. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nature Methods. 5, 865-867 (2008).
- Lewis, J. A., Fleming, J. T. Basic culture methods. Methods in Cell Biology. 48, 3-29 (1995).
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics. 77, 71-94 (1974).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved