A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Concentric Gel System to Study the Biophysical Role of Matrix Microenvironment on 3D Cell Migration
In This Article
Summary
The mechanical properties and microstructure of the extracellular matrix strongly affect 3D migration of cells. An in vitro method to study the spatiotemporal cell migration behavior in biophysically variable environments, at both population and individual cell levels, is described.
Abstract
The ability of cells to migrate is crucial in a wide variety of cell functions throughout life from embryonic development and wound healing to tumor and cancer metastasis. Despite intense research efforts, the basic biochemical and biophysical principles of cell migration are still not fully understood, especially in the physiologically relevant three-dimensional (3D) microenvironments. Here, we describe an in vitro assay designed to allow quantitative examination of 3D cell migration behaviors. The method exploits the cell’s mechanosensing ability and propensity to migrate into previously unoccupied extracellular matrix (ECM). We use the invasion of highly invasive breast cancer cells, MDA-MB-231, in collagen gels as a model system. The spread of cell population and the migration dynamics of individual cells over weeks of culture can be monitored using live-cell imaging and analyzed to extract spatiotemporally-resolved data. Furthermore, the method is easily adaptable for diverse extracellular matrices, thus offering a simple yet powerful way to investigate the role of biophysical factors in the microenvironment on cell migration.
Introduction
Migration of cells plays a key role in various physiological responses such as embryonic development, haemostasis, and immune response as well as in pathological processes such as vascular diseases, inflammation, and cancer1. Dissecting the biochemical and biophysical factors underlying cell migration is therefore fundamentally important not only to understand the basic principles of cellular functions, but also to advance various biomedical applications, such as in tissue engineering, anti-metastasis and anti-inflammatory drug development. Since in vivo observation is technically challenging, a lot of efforts has been focused on in vitro recapitulation of cell migration.
In vitro methods to study cell migration have largely been designed for assays on two dimensional (2D) surfaces, most notably the scratch or wound healing assay2. Such assays offer simple experimental setup, easy live-cell imaging, and provide useful insights into various biochemical mechanisms underlying cell migration. However, these assays do not account for extracellular matrix (ECM) architecture and remodeling, which are critical aspects in understanding in vivo migration. Recently, it has been increasingly appreciated that a 3D culture model, often in collagen-based matrices3, provides a platform that better resembles the in vivo situation. Indeed, cells exhibit migrational dynamics that are distinct from those on 2D surfaces, especially due to the different dimensionality of the environment4. Moreover, the biophysical and mechanical properties of the matrix sensitively affect cell migration5, including in the context of tumor cell invasion6.
Here, we present a method to study 3D cell migration behavior in ECM with biophysical properties that can be easily varied with preparation conditions. The cells are seeded in an “inner gel” and are allowed to escape into and invade the initially acellular “outer gel”. The method relies on the cell’s ability to recognize the presence of, and propensity to migrate into, cell-free regions in the outer gel, which is closely linked to cell mechanosensing7. In this study, we employ collagen networks as the ECMs invaded by highly invasive breast cancer cells, MD-MBA-231. The mechanical properties and microstructure of both the inner and outer gels can be tuned8 and characterized9 to achieve physiologically relevant conditions. Reconstruction and analysis of the cell tracks allow detailed quantitative examination of the spatiotemporal migration behavior at both population level and individual cell level. Importantly, the setup of the concentric gel system mimics the in vivo tissue topology faced by migrating cells, especially invading cancer cells, thus offering important insights into the physical mechanisms of cell migration and metastasis.
Protocol
1. Cell Harvesting
- Obtain MD-MBA-231 cells from the 37 °C, 5% CO2 incubator. Detach cells from tissue culture plate using 0.5% Trypsin-EDTA solution. Use 1 ml of Trypsin-EDTA solution for cell cultured in a T25 flask.
- Pellet cells in a 15 ml conical tube by centrifugation at 200 × g for 4 min, aspirate the supernatant, and resuspend cells in 5 ml of culture media.
- Count cell density, ρ, using a hemocytometer.
Note: To prepare the cell-seeded inner gel, the cell suspension will later on be diluted 10× to reach the final cell seeding density. Therefore, 10× concentrated cell suspension is required. - Calculate the volume of medium required to achieve a 10× cell concentration:
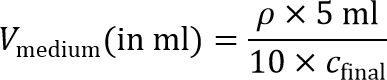
Note: A final cell seeding density, cfinal, of around 2 × 106 cells/ml is recommended for MD-MBA-231 cells and is used in this protocol. Other seeding densities can also be explored for other cell types. - Pellet cells one more time in a 15 ml conical tube by centrifugation at 200 × g for 4 min, and aspirate the supernatant.
- Resuspend the cells in the required amount (Vmedium calculated in step 1.3) of serum-free cell culture medium thoroughly to minimize cell clumping.
Note: Phenol red is auto-fluorescent, and can interfere with fluorescence/reflectance imaging. Use of phenol red-free medium may be considered to achieve best image quality.
2. Preparation of Collagen Solutions
- Obtain the stock collagen solution, 10× PBS buffer, Milli-Q H2O, 0.1 M NaOH, and several microcentrifuge tubes. Keep all on ice to prevent premature collagen polymerization, and maintain sterile condition.
- Equilibrate a sterile glass-bottom dish by pre-warming it in the 37 °C incubator.
Note: All volumes in this protocol have been optimized for a glass-bottom dish with 12 mm well. If other dish type is used, adjust the volumes correspondingly. - Calculate the required volume needed to prepare 50 µl of 2.4 mg/ml collagen solution for the inner gel (solution I) based on the collagen stock concentration.
Note: Other collagen concentrations for the inner gel can also be used. - In a sterile environment (typically a biosafety hood), slowly add 5 µl of 10× PBS buffer to the required amount of collagen stock solution (calculated in step 2.3) with gentle swirling. Take care to avoid air bubble formation.
- Adjust the pH of the mixture to 7.4 using 0.1 M NaOH using calibrated pH-meter. As a rough guide, use about 5 µl to bring the pH close to 7.4 (the amount varies depending on the stock concentration and pH).
Note: Note that the volumes involved in this step is too small for the use of standard pH meter. Use one of the following tricks:- Prepare collagen solutions for multiple samples. Adjust the pH in bulk using standard pH meter and distribute the collagen solutions across the samples.
- Alternatively, adjust the pH of a collagen solution in larger volume (i.e., volume that allow the use of standard pH meter). Note the amount of NaOH needed to bring the pH to the final pH. Scale down the volumes and use the appropriate amount of NaOH for the experiment. Confirm the pH value using Litmus paper.
- Otherwise, use a micro pH electrode to more accurately adjust the pH of small quantities.
- Bring the solution to a volume of 45 µl using H2O. Perform all steps on ice to prevent premature collagen polymerization.
3. Formation of Concentric Gel Culture
- Take the pre-warmed glass-bottom dish (see step 2.2) from the incubator.
- Add 5 µl of the 10× concentrated cell suspension (prepared in step 1.5) to solution I. Resuspend thoroughly. Take care to avoid air bubble formation. The mixture now has a volume of 50 µl and contains the final collagen concentration (2.4 mg/ml) and cell density (cfinal = 2 × 106 cells/ml).
- Add 20 µl of the cell-containing solution I slowly to the center of the well, so that it forms a dome-shaped droplet (Figure 1A). Take care to avoid bubble formation at this step. If a bubble forms, carefully but quickly try to either rupture it or suck it out using a pipette tip. Gently place the dish back in the incubator to let the inner gel polymerize for 45 min.
- Prepare solution O (for the outer, acellular collagen gel) about 15 min before the end of this incubation step.
Note: The outer gel condition can be varied in terms of collagen concentration and polymerization pH to obtain networks with different microstructures10. In this protocol, focus on a 1.5 – 4.0 mg/ml collagen gel polymerized at a pH of 7.4.- Based on the collagen stock concentration, calculate the required volume needed to prepare 200 µl of collagen solution O at the final concentration.
- Add 20 µl of 10× PBS buffer slowly to the required amount of collagen stock solution (calculated in step 3.4) with gentle swirling. Adjust the pH of the mixture to the final pH using 0.1 M NaOH with the use of calibrated pH-meter. See note to step 2.5 regarding pH adjustment.
- Bring the solution to a final volume of 200 µl using H2O. Perform all steps on ice to prevent premature collagen polymerization.
- Take the dish from the incubator after 45 min polymerization of the inner gel (see step 3.3). Gently add 180 µl of solution O on top of the inner gel, so that the solution completely covers the inner gel and fills up the well (Figure 1B).
- Perform this step carefully without stirring the solution, which can lead to non-uniform fiber orientations in the outer gel. Take care to avoid touching the inner gel with a pipette tip, and to avoid the formation of bubbles or air pockets. If a bubble forms, carefully but quickly try to either rupture it or suck it out using a pipette tip. Gently place the dish back in the incubator to let the outer gel polymerize.
- Take the dish from the incubator after 45 min of polymerization. The gel should already be fairly solidified at this point, although it can still detach from the bottom surface if handled roughly.
- Gently pour 2 ml of warmed cell culture medium to the dish (Figure 1C). Make sure that the gel is completely submerged in the medium. Refresh the medium every 2 – 3 days throughout the duration of culture.
4. Live-cell Imaging
- Perform imaging using an inverted confocal microscope equipped with long-term live-cell imaging capability. Include a built-in incubation chamber with a temperature (37 °C) and CO2 (5%) control. Switch on the microscope and warm up the stage at least 1 hr before starting the experiment.
Note: Use objective lens with long working distance to optimize the observation and localization of cells in the 3D gels. - Incubate the gel in medium containing 5 µl of fluorescent cell tracker dye for 30 min to allow accurate localization of the cells in the 3D system. Subsequently, remove unbound dye by washing three times with 1× PBS. Afterwards, add cell culture medium to the dish.
- Take the dish from the incubator and place it on the microscope stage (Figure 1D).
Note: Live cell imaging can start in principle right after the polymerization of the outer gel. However at this point, the cells in the inner gel have not spread yet. To examine the migration of the first cells that have invaded the outer gel, live-cell imaging can start 24 hr (depending on the cell type) after the initiation of culture. As a rough guide, around 12 - 14 days of culture are needed for most of the cells in the inner gel to enter the outer gel. - Select Volumes of View (VoV’s) in regions in the outer gel surrounding the inner gel. After 24 hr of incubation, the cell population will have spread, crossed the interface between the inner and outer gels, and started to invade the outer gel.
- For the VoV’s, include the gel regions immediately next to the gel interface, intermediate regions, and regions close to the outskirt of the outer gel7. Exclude regions closer than 50 µm from the bottom and side surfaces, as well as from the top of the gel, to avoid possible edge effects. Each VoV typically measures 647 × 647 × 100 µm3 (in x, y, and z directions, respectively), with 5 µm interval in the z-stack.
- Ensure imaging modes, channels/filters, exposure times, and image resolutions are correctly selected. For label-free imaging of the collagen network, use confocal reflectance microscopy simultaneously during the time-lapse live-cell imaging.
- Take sample images, plot the intensity histograms, and adjust the gains and offsets to observe sufficient signal and avoid saturation by ensuring that the histogram lies between zero and the maximum intensity. Do not change these settings any more throughout the duration of the experiment.
- Take time-lapse images of the selected VoV’s, with a time interval, Δt, of 10 min for 8 hrs (or longer if necessary).
5. Cell Tracking and Data Analysis
- Carry out quantitative image post-processing from the z-stack images using appropriate image processing software.
- Segment the time-lapse images to automatically select the 3D cell positions in (x,y,z,t).
- For every frame, manually check the localization accuracy and remove false positives due to cell debris and cellular protrusions that may have been mistaken for cells. Remove proliferative cells from the analysis and split overlapping or connected cells into distinct objects.
- Generate 3D time-lapse cell tracks from the cell coordinates (x,y,z,t) obtained in the previous step by linking the location of each cell in the time sequence.
- Eliminate random and system noise by removing tracks shorter than a threshold track length (typically 20 min).
- Correct for sample drift by subtracting the overall net displacements from the tracks if necessary.
- Calculate cell displacement,
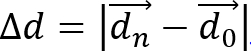 , and cell migration distance,
, and cell migration distance, 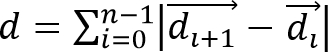 , from the observed cell trajectories, where is a vector representing the 3D location of a cell at time and is the total number of time points.
, from the observed cell trajectories, where is a vector representing the 3D location of a cell at time and is the total number of time points.
- Calculate cell speed as S = d/(n • Δt), where Δt is the time interval between frames. Calculate cell migration directionality (or persistence) using P = Δd/d. This simple measure of persistence implies that for P = 0 the net displacement is zero and for P = 1 the trajectory is a straight directional line.
Results
The concentric gel assay presented here was performed using highly invasive breast cancer cells, MDA-MB-231, with 2.4 mg/ml inner collagen gel and a cell seeding density of = 2 × 106 cells/ml, as an example. As shown in Figure 2, typically after a few days of culture, the cells breached the inner–outer gel interface and started to invade the outer gel. The cell population spread predominantly radially outwards.
The polymerization conditions of the outer g...
Discussion
In this protocol we describe an in vitro assay to study the 3D migrational behavior of cells in matrix environments that topologically resemble ECMs encountered in vivo. There are three main strengths of this assay as compared to other currently available methods. First, this assay allows one to simultaneously examine the cell migration mechanisms at both population level and individual cell level. This opens up possibilities of studying collective cell migration13, which has to date been lar...
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank W. Sun and K. Jansen for the critical discussions, and acknowledge support by the Nano Biomechanics Lab at the National University of Singapore. N.A.K. acknowledges support by a Marie Curie IIF Fellowship.
Materials
| Name | Company | Catalog Number | Comments |
| Cell culture incubator | Fisher Scientific Pte Ltd | Model: 371, S/No 318854-6055 | |
| Confocal microscope | Nikon A1R | Inverted confocal laser scanning microscope equipped with incubator chamber | |
| Dulbecco's Modified Eagle's Medium (DMEM) | Life Technologies | 11965-092 | |
| Fetal Bovine Serum (FBS) | Life Technologies | 10082-147 | |
| Fluorescent CellTracker dye CMTMR | Life Technologies | C2927 | |
| Glass-bottom dish | IWAKI Cell Biology | 3931-035 | 35 mm diameter dish with 12 mm diameter glass-bottom well |
| Hemocytometer | iN CYTO | DHC-N01 (Neubauer Improved) | |
| Microprocessor pH meter | Hanna Instruments | pH 211 | |
| Nutragen Collagen | Advanced BioMatrix | #5010-D | Acid-solubilized bovine collagen type I (stock pH ~ 2) |
| Objective lens | Nikon | CFI Super Plan Fluor ELWD ADM 20XC, W.D. 8.2-6.9mm, NA 0.45. | |
| Penicillin-Streptomycin | Life Technologies | 15140-122 | |
| pH meter | Sartorius | S/No 29153352 | Basic pH Meter PB-11 |
| Trypsin-EDTA | Life Technologies | 15400-054 |
References
- Horwitz, R., Webb, D. Cell migration. Curr Biol. 13 (19), R756-R759 (2003).
- Liang, C. C., Park, A. Y., Guan, J. L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2 (2), 329-333 (2007).
- Provenzano, P. P., Eliceiri, K. W., Inman, D. R., Keely, P. J. Engineering three-dimensional collagen matrices to provide contact guidance during 3D cell migration. Curr. Prot. Cell Biol. 10, 10-17 (2010).
- Friedl, P., Sahai, E., Weiss, S., Yamada, K. M. New dimensions in cell migration. Nat. Rev. Mol. Cell Biol. 13 (11), 743-747 (2012).
- Grinnell, F., Petroll, W. M. Cell motility and mechanics in three-dimensional collagen matrices. Annu. Rev. Cell. Dev. Biol. 26, 335-361 (2010).
- Wirtz, D., Konstantopoulos, K., Searson, P. C. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer. 11 (7), 512-522 (2011).
- Sun, W., Kurniawan, N. A., Kumar, A. P., Rajagopalan, R., Lim, C. T. Effects of migrating cell-induced matrix reorganization on 3D cancer cell migration. Cell. Mol. Bioeng. 7 (2), 205-217 (2014).
- Achilli, M., Mantovani, D. Tailoring mechanical properties of collagen-based scaffolds for vascular tissue engineering: the effects of pH, temperature and ionic strength on gelation. Polymers. 2 (4), 664-680 (2010).
- Kurniawan, N. A., Wong, L. H., Rajagopalan, R. Early stiffening and softening of collagen: interplay of deformation mechanisms in biopolymer networks. Biomacromolecules. 13 (3), 691-698 (2012).
- Sun, W., Lim, C. T., Kurniawan, N. A. Mechanistic adaptability of cancer cells strongly affects anti-migratory drug efficacy. J. R. Soc. Interface. 11 (99), 20140638 (2014).
- Guzman, A., Ziperstein, M. J., Kaufman, L. J. The effect of fibrillar matrix architecture on tumor cell invasion of physically challenging environments. Biomaterials. 35 (25), 6954-6963 (2014).
- Wolf, K., et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201 (7), 1069-1084 (2013).
- Friedl, P., Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10 (7), 445-457 (2009).
- Vedula, S. R. K., Ravasio, A., Lim, C. T., Ladoux, B. Collective cell migration: a mechanistic perspective. Physiology. 28 (6), 370-379 (2013).
- Petrie, R. J., Doyle, A. D., Yamada, K. M. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10 (8), 538-549 (2009).
- Miron-Mendoza, M., Seemann, J., Grinnell, F. The differential regulation of cell motile activity through matrix stiffness and porosity in three dimensional collagen matrices. Biomaterials. 31 (25), 6425-6435 (2010).
- Mouw, J. K., et al. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat. Med. 20 (4), 360-367 (2014).
- Wolf, K., et al. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 20 (8), 931-941 (2009).
- Wong, L. H., Kurniawan, N. A., Too, H. -. P., Rajagopalan, R. Spatially resolved microrheology of heterogeneous biopolymer hydrogels using covalently bound microspheres. Biomech. Model. Mechanobiol. 13 (4), 839-849 (2014).
- Gupton, S. L., Waterman-Storer, C. M. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 125 (7), 1361-1374 (2006).
- Provenzano, P. P., Inman, D. R., Eliceiri, K. W., Trier, S. M., Keely, P. J. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys. J. 95 (11), 5374-5384 (2008).
- Wolf, K., et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9 (8), 893-904 (2007).
- Jansen, K. A., Bacabac, R. G., Piechocka, I. K., Koenderink, G. H. Cells actively stiffen fibrin networks by generating contractile stress. Biophys. J. 105 (10), 2240-2251 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved