A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Interactions with and Membrane Permeabilization of Brain Mitochondria by Amyloid Fibrils
* These authors contributed equally
In This Article
Summary
Provided here is a protocol for investigating the interactions between native form, prefibrillar, and mature amyloid fibrils of different peptides and proteins with mitochondria isolated from different tissues and various areas of the brain.
Abstract
A growing body of evidence indicates that membrane permeabilization, including internal membranes such as mitochondria, is a common feature and primary mechanism of amyloid aggregate-induced toxicity in neurodegenerative diseases. However, most reports describing the mechanisms of membrane disruption are based on phospholipid model systems, and studies directly targeting events occurring at the level of biological membranes are rare. Described here is a model for studying the mechanisms of amyloid toxicity at the membrane level. For mitochondrial isolation, density gradient medium is used to obtain preparations with minimal myelin contamination. After mitochondrial membrane integrity confirmation, the interaction of amyloid fibrils arising from α-synuclein, bovine insulin, and hen egg white lysozyme (HEWL) with rat brain mitochondria, as an in vitro biological model, is investigated. The results demonstrate that treatment of brain mitochondria with fibrillar assemblies can cause different degrees of membrane permeabilization and ROS content enhancement. This indicates structure-dependent interactions between amyloid fibrils and mitochondrial membrane. It is suggested that biophysical properties of amyloid fibrils and their specific binding to mitochondrial membranes may provide explanations for some of these observations.
Introduction
Amyloid-related disorders, known as amyloidoses, constitute a large group of diseases defined by the appearance of insoluble protein deposits in different tissues and organs1,2. Among them, neurodegenerative disorders are the most frequently forms in which protein aggregates appear in the central or peripheral nervous system2. Although a number of mechanisms have been proposed to be involved in the toxicity of amyloid aggregates3, a growing body of evidence points to cell membrane disruption and permeabilization as the primary mechanism of amyloid pathology4,5. In addition to plasma membrane, internal organelles (i.e., mitochondria) may also be affected.
Interestingly, emerging evidence suggests that mitochondrial dysfunction plays a critical role in the pathogenesis of neurodegenerative disorders, including Alzheimer’s and Parkinson’s diseases6,7. In accordance with this issue, numerous reports have indicated binding and accumulation of amyloid β-peptide, α-synuclein, Huntingtin, and ALS-linked mutant SOD1 proteins to mitochondria8,9,10,11. The mechanism of membrane permeabilization by amyloid aggregates is thought to occur either through formation of discrete channels (pores) and/or through a nonspecific detergent-like mechanism5,12,13. It is noteworthy that most of these conclusions have been based on reports involving phospholipid model systems, and studies directly targeting the events occurring in biological membranes are rare. Clearly, these artificial lipid bilayers do not necessarily reflect the intrinsic properties of biological membranes, including those of mitochondria, which are heterogeneous structures and composed of a wide variety of phospholipids and proteins.
In the present study, mitochondria isolated from rat brains are used as an in vitro biological model to examine the destructive effects of amyloid fibrils arising from α-synuclein (as an amyloidogenic protein), bovine insulin (as a model peptide showing significant structural homology with human insulin involved in injection-localized amyloidosis), and hen egg white lysozyme (HEWL; as a common model protein for study of amyloid aggregation). The interactions and possible damage of mitochondrial membranes induced by amyloid fibrils are then investigated by observing the release of mitochondrial malate dehydrogenase (MDH) (located in the mitochondrial matrix) and mitochondria reactive oxygen species (ROS) enhancement.
Protocol
All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) of Medical Sciences of Tehran University. Maximal efforts were made to minimize suffering and detrimental effects to the rats by sharpening the guillotine blades and applying resolute and swift movements of the blade.
1. Brain homogenization and mitochondrial isolation
NOTE: All reagents for mitochondrial isolation were prepared according to Sims and Anderson14.
- Preparation of buffers for mitochondrial isolation
- Prepare a 100 mM Tris-HCl solution: weigh 0.605 g of Tris-HCl and dissolve it in approximately 40 mL of deionized water (DW) in a beaker. Transfer the solution to a 50 mL volumetric flask and increase the final volume to 50 mL by adding DW.
- Prepare a 10 mM EDTA solution: weigh 0.202 g of EDTA and dissolve it in approximately 40 mL of DW in a beaker. Transfer the solution to a 50 mL volumetric flask and increase the final volume to 50 mL by adding DW.
NOTE: Both solutions can be stored at 4 °C for at least 4 weeks. - Prepare a Tris-HCl/EDTA/sucrose stock solution: add 30 mL of 100 mM Tris-HCl and 30 mL of 10 mM EDTA in a beaker. Weigh 32.86 g of sucrose and add it to the beaker while stirring with a magnetic stirrer. When the sucrose is dissolved, transfer the solution to a 100 mL volumetric flask and increase the final volume to 100 mL by adding DW.
NOTE: This solution can be store at 4 °C for up to 3 days. - Prepare an isolation buffer (IB): add 100 mL of Tris-HCl/EDTA/sucrose stock to approximately 150 mL of DW. Adjust the pH to 7.4 by adding 0.1 M HCl. Increase the final volume to 300 mL by adding DW.
- Prepare 15% (v/v) density gradient medium with IB by adding 1.5 mL of density gradient medium to 8.5 mL of cold IB.
- Prepare 10 mg/mL bovine serum albumin (BSA) solution: weigh 20 mg of fatty acid-free BSA and dissolve it in approximately 1.5 mL DW in a 2 mL vial. Increase the final volume to 2 mL by adding DW and store it on ice until use.
NOTE: Freshly prepare the solutions from steps 1.1.4–1.1.6 on the day of mitochondrial isolation and store them on ice until use.
- Isolation of rat brain
NOTE: Decapitation and brain removal of male rats (150–200 g) were performed according to Sims and Anderson14.- Put a 30 mL beaker in an ice bucket and add 10 mL of cold IB to the beaker.
- Decapitate the rat with a small animal guillotine and remove the brain from the skull within 1 min of decapitation to limit deterioration of mitochondrial properties.
- Rapidly transfer the brain to the beaker containing cold IB.
- Brain homogenization and preparation of mitochondria
NOTE: Mitochondrial fractions were isolated according to the protocol described by Sims and Anderson14, with some modifications as described previously15. It is important to work quickly and keep everything on ice throughout the procedure.- Wash the tissue 2x with 30 mL of IB, transfer to a beaker containing cold IB, and finely mince the brain with scissors.
- Add 10 volumes of pre-cooled IB (about 10 mL of IB per rat brain).
- Transfer the tissue suspension to a 20 mL cold Dounce homogenizer.
- Homogenize the tissue pieces using nine up-and-down strokes with a motorized pestle.
NOTE: Leave the mixture on ice for approximately 30 s after each set of three homogenization strokes to ensure that the homogenate remains cold. - Transfer the homogenate to a pre-chilled 10 mL centrifuge tube and centrifuge at 1,300x g and 4 °C for 3 min.
- Carefully decant the supernatant and transfer it to a pre-chilled 10 mL centrifuge tube and centrifuge at 21,000x g at 4 °C for 10 min.
- Discard the supernatant and resuspend the pellet in a cold 15% density gradient medium solution (5 mL for each brain) by gently stirring the mixture with a pipette.
- Centrifuge in a fixed-angle rotor at 30,700x g at 4 °C for 5 min using slow acceleration (45 s from 0 rpm to 500 rpm followed by normal acceleration) and deceleration (no brakes). This should produce two distinct bands of material (Figure 1A, left).
- Using a Pasteur pipette, remove the sharp band of material accumulated at the top of the gradient, which mostly contains myelin. Then, remove the density gradient medium solution overlying the material (in band 2) as much as possible without losing any of the enriched mitochondrial fraction in band 2.
NOTE: The band 2 contains both the synaptic and non-synaptic mitochondria. - Add 8 mL of IB to the mitochondrial fraction while gently stirring the mixture with a pipette.
- Centrifuge at 16,700x g at 4 °C for 10 min and carefully remove the supernatant, leaving the bottom loose pellet undisturbed.
- Add 1 mL of 10 mg/mL fatty acid-free BSA to the centrifuge tube while gently stirring the mixture with the tip of a pipette. Increase the final volume to 5 mL per brain by adding IB.
- Centrifuge at 6,900x g at 4 °C for 10 min, which should produce a firm pellet (Figure 1A, right).
- Decant the supernatant and gently resuspend the mitochondrial pellet in IB, aliquot them into 0.5 mL tubes, and store in liquid nitrogen until use.
2. Protein concentration determination
NOTE: Protein concentration is measured using the method of Lowry et al.16.
- Preparation of solutions for Lowry assay
- Prepare solution A: weigh 0.4 g of NaOH and 2 g of Na2CO3 and dissolve in 80 mL of DW, then transfer the solution to a 100 mL volumetric flask and increase the volume to 100 mL by adding DW.
- Prepare solution B: weigh 0.1 g of potassium sodium tartrate and 0.05 g of CuSO4 and dissolve in 8 mL of DW, then transfer the solution to a 10 mL volumetric flask and increase the volume to 10 mL by adding DW.
NOTE: Both solutions A and B can remain stable at 4 °C for up to 6 months. - Prepare solution C: Add 0.5 mL of folin solution to 7.5 mL of DW.
NOTE: Prepare solution C fresh and keep away from light until use. - Prepare BSA standards with final concentrations of 0, 20, 40, 60, 80, 100, and 120 µg/mL by combining 0, 20, 40, 60, 80, 100, and 120 µL of 1 mg/mL BSA, respectively, with enough DW to make 1000 µL of solution.
- Protein concentration measurement
NOTE: For greater accuracy, run this step in triplicate.- Add 50 µL of standard solutions and mitochondrial homogenate to each well of a 96 well plate, followed by the addition of 45 µL of solution A. Then, incubate the plate for 10 min in a warm water bath set at 50 °C.
- Add 5 µL of solution B and incubate the plate for 10 min in dark at room temperature (RT).
- Add 150 µL of solution C and incubate the plate for 10 min in a warm water bath set at 50 °C.
- Load the plate into a plate reader and record the absorbance values for standards and mitochondrial suspension at 650 nm. Then, using a calibration curve, calculate the protein content of the mitochondria.
3. Mitochondrial membrane integrity determination
NOTE: Mitochondrial membrane integrity is confirmed by measuring malate dehydrogenase (MDH) activity in isolated mitochondria before and after membrane disruption by Triton X-100.
- Dilute mitochondrial homogenate to 1 mg/mL with cold IB and placed in two 1.5 mL tubes (typically 195 μL of mitochondria per tube).
- Add 5 µL of 20% (v/v) Triton X-100 (diluted with DW) to one tube (as a positive control for maximum enzyme activity) and 5 µL of DW to another tube (for control) followed by mixing with a stirrer.
- Incubate the tubes for 10 min in a warm water bath set at 30 °C.
- Pellet mitochondria by centrifugation of tubes in a fixed-angle rotor at 16,000 x g and 4 °C for 15 min.
- Carefully collect the resulting supernatants for assaying the activity of mitochondrial MDH using a standard spectrophotometric assay described in the following section.
- Calculate the integrity of mitochondrial membrane as follows:
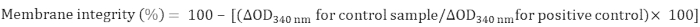
4. MDH activity determination
NOTE: MDH activity was measured spectrophotometrically as described by Sottocasa et al.17.
- Preparation of solutions for MDH activity assay
- Prepare a 50 mM Tris-HCl solution (pH = 7.5): prepare a 7.9 mg/mL solution in DW using Tris-HCl, and adjust the pH to 7.5 at 25 °C with 1.0 M NaOH.
- Prepare a 50 mM oxaloacetate solution: prepare a 6.6 mg/mL solution using oxaloacetate in Tris-HCl.
NOTE: This solution is not stable once in solution and should be prepared immediately prior to use. - Prepare a 10 mM β-NADH solution: prepare a 7.81 mg/mL solution using β-NADH in Tris-HCl.
- MDH activity determination
NOTE: Final assay concentrations in a 1.0 mL reaction mixture are 50 mM Tris-HCl, 5 mM oxaloacetate, and 0.1 mM β-NADH.- Set the spectrophotometer to 25 °C and 340 nm. Pipette 890 µL Tris-HCl buffer, 100 µL of oxaloacetate solution, and 10 µL of NADH solution to a blank cuvette and 880 µL Tris-HCl buffer, 100 µL of oxaloacetate solution, and 10 µL of NADH solution into a sample cuvette.
- Incubate cuvettes in the spectrophotometer for 3–4 min and reference against blank.
- Then add 10 µL of mitochondrial homogenate (1 mg/mL) to the sample cuvette, immediately mix by inversion, and record decreases in absorbance due to NADH oxidation at 340 nm for 1 min.
5. In vitro α-synuclein, bovine insulin, and HEWL fibril formation
- Protein preparation
- α-synuclein:
NOTE: Expression and purification of recombinant α-synuclein is performed as described by Hoyer et al.18 with some modifications, and the purity of α-synuclein is confirmed by SDS-PAGE.- Dialyze purified α-synuclein overnight against phosphate-buffered saline (PBS).
- Determine the protein concentration using an extinction coefficient of 5600 M-1 cm-1 at 275 nm19.
- Aliquots protein in 1.5 mL tubes and store at -80 °C until use.
- Bovine insulin and HEWL:
- Provide both bovine insulin and HEWL.
- Dissolve each protein in 50 mM glycine buffer (pH = 1.6; adjust the pH with HCl).
- Determine bovine insulin and HEWL concentration using an extinction coefficient of 1.0 and 2.63 for 1.0 mg/mL at 276 nm and 280 nm, respectively20,21.
- α-synuclein:
- Amyloid fibrillation induction
- Preparation of solutions for amyloid fibrillation:
- Prepare thioflavin T (ThT) buffer: weigh 0.3 g of NaH2PO4 and dissolve it in 80 mL of DW, adjust the pH to 6.5, and increase the volume to 100 mL by adding DW.
- Prepare ThT stock solution (5 mM): weigh 1.6 mg of ThT and dissolve it in 1 mL of ThT buffer, pass through a 0.22 μm filter paper, and keep away from light at 4 °C until use.
NOTE: This solution can be store at 4 °C for up to 4 weeks. - Prepare ThT solution (1 mM): add 200 µL of ThT stock solution (5 mM) to 800 µL of ThT buffer.
NOTE: This solution can be stored at 4 °C for up to 1 week.
- In vitro α-synuclein amyloid fibril formation:
- Add aliquots (294 µL) of protein solution (200 μM) and 6 µL of ThT solution (1 mM) to 1.5 mL tubes followed by stirring.
- Incubate the tubes in a thermomixer at 37 °C under constant stirring at 800 rpm for 4 days.
- Take aliquots (10 µL) of incubated samples after regular time intervals and add 490 µL of ThT buffer, mixed thoroughly, and incubate for 5 min at RT.
- Set excitation and emission slit widths as 5 nm and 10 nm, respectively, and measure ThT fluorescence by excitation at 440 nm and emission at 485 nm using a fluorescence spectrophotometer.
- In vitro bovine insulin amyloid fibril formation:
- Add 637 µL of protein solution (250 µM) to 1.5 mL tube. Then, add 13 µL of ThT solution (1 mM) followed by stirring.
- Add aliquots (200 µL) of protein solution (250 µM) containing 20 µM ThT to each well of a clear-bottomed 96 well plate and seal the plate with crystal clear sealing tape.
- Load the plate into a fluorescence plate reader and incubate at 57 °C without agitation.
- Measure ThT fluorescence at 30 min intervals, with excitation at 440 nm and emission at 485 nm, for 12 h.
NOTE: Shake the plate for 5 s before each measurement.
- In vitro HEWL amyloid fibril formation:
- Add aliquots (200 µL) of protein solution )1 mM) containing 20 µM ThT to each well of a clear-bottomed 96 well plate and seal the plate with crystal clear sealing tape.
- Load the plate into a fluorescence plate reader and incubate at 57 °C without agitation.
- Measure ThT fluorescence at 2 h intervals, with excitation at 440 nm and emission at 485 nm, for 4 days.
NOTE: For all three proteins, amyloid fibril formation is confirmed by atomic force microscopy (Figure 2B).
- Preparation of solutions for amyloid fibrillation:
6. Treatment of mitochondria with amyloid fibrils, MDH release assay, and ROS measurement
- Incubation of isolated mitochondria with amyloid fibrils
- Turn on centrifuge and warm water bath and set to 4 °C and 30 °C, respectively.
- Using IF, dilute mitochondrial homogenate to a final concentration of 1 mg/mL.
- Prepare two series of 1.5 mL tubes containing mitochondrial homogenates (one series for MDH release assay and another for mitochondrial ROS measurement).
- Add aliquots of fresh or amyloid fibrils of α-synuclein, bovine insulin, or HEWL (at final concentrations of 5 µM, 10 µM, 20 µM, and 25 µM; use PBS or glycine buffer as a control) to mitochondrial homogenate (final volume = 200 µL) (see Table 1, Table 2, and Table 3) followed by gently stirring the solution with a pipette.
NOTE: For the MDH release assay, use Triton X-100 (at a final concentration of 0.5% [v/v]) as a positive control for maximum enzyme release. - Incubate tubes containing mitochondrial suspensions for 30 min in warm water bath set to 30 °C.
- Measure mitochondrial MDH release and ROS content as outlined in sections 6.2 and 6.3.
- Mitochondrial MDH release assay
- Centrifuge the incubated mitochondrial homogenates at 16,000 x g for 15 min, then carefully collect the resulting supernatants for assaying the activity of mitochondrial MDH as described in section 4.
- Calculate the release of MDH as a fraction of the maximum effect (Triton X-100) as follows:
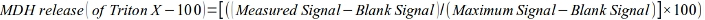
- Mitochondrial ROS measurement
NOTE: Mitochondrial ROS content is determined with the oxidation sensitive fluorogenic precursor dihydrodichlorocarboxyfluorescein diacetate (DCFDA)22.- Prepare solutions for mitochondrial ROS measurement. Prepare a 50 µM DCFDA solution by dissolving in methanol (prepare fresh) and a 200 mM succinate solution by dissolving in DW.
- Pipette 191 µL of incubated mitochondrial homogenate to each well of a 96 well plate and add 4 µL of 50 µM DCFDA (1 µM final concentration) and 5 µL of 200 mM succinate (5 mM final concentration).
- Incubate the plate for 30 min in a warm water bath set at 30 °C while gently stirring.
- Load the plate into a fluorescence plate reader and measure fluorescence intensity, with excitation at 485 nm and emission at 530 nm.
7. Statistical analysis
- Perform all experiments at least 2x or 3x with triplicate assays and conduct appropriate statistical tests. Here, the results are presented as mean ± SD, and a Student's paired t-test was utilized to calculate statistical significance. P-values less than 0.01 and 0.05 were considered statistically significant (*p < 0.05; **p < 0.01).
Results
The protocol describes a model for studying the interactions of amyloid fibril with rat brain mitochondria as an in vitro biological model. For mitochondrial preparation, 15% (v/v) density gradient medium was used to remove myelin as major contamination of brain tissue14. As shown in Figure 1A, centrifugation at 30,700 x g produced two distinct bands of material, myelin (as the major component of band 1) and band 2, which...
Discussion
A wealth of experimental results supports the hypothesis that the cytotoxicity of fibrillar aggregates is significantly associated with their ability to interact with and permeabilize biological membranes4,5. However, most of the data are based on artificial lipid bilayers that do not necessarily reflect the intrinsic properties of biological membranes, which are heterogeneous structures with a wide variety of phospholipids and proteins. Here, using brain mitocho...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants from the Research Council of the Institute for Advanced Studies in Basic Sciences (IASBS), Zanjan, Iran.
Materials
| Name | Company | Catalog Number | Comments |
| 2′,7′-Dichlorodihydrofluorescein diacetate | Sigma | 35845 | |
| Ammonium sulfate | Merck | 1012171000 | |
| Black 96-well plate | Corning | ||
| Black Clear-bottomed 96-well plate | Corning | ||
| Bovine insulin | Sigma | I6634 | |
| Bovine Serum Albumin (BSA) | Sigma | A2153 | |
| BSA essentially fatty acid-free | Sigma | A6003 | |
| Centrifuge | Sigma | ||
| Crystal clear sealing tape | Corning | ||
| CuSO4 | Sigma | 451657 | |
| Dialysis bag (cut off 2 KDa) | Sigma | D2272 | |
| Dounce homogenizer | Potter Elvehjem | ||
| EDTA | Sigma | E9884 | |
| Fluorescence plate reader | BioTek | ||
| Fluorescence spectrophotometer | Cary Eclipse VARIAN | ||
| Folin | Merck | F9252 | |
| Glycine | Sigma | G7126 | |
| Guillotine | Made in Iran | ||
| HCl | Merck | H1758 | |
| Hen Egg White Lysozyme (HEWL) | Sigma | L6876 | |
| Na2CO3 | Sigma | S7795 | |
| NaH2PO4 | Sigma | S7907 | |
| NaOH | Merck | S8045 | |
| Oxaloacetate | Sigma | O4126 | |
| Percoll | GE Healthcare | ||
| Phosphate Buffer Saline (PBS) | Sigma | CS0030 | |
| PMSF | Sigma | P7626 | |
| Potassium sodium tartrate | Sigma | 217255 | |
| Quartz cuvette | Sigma | ||
| Spectrophotometer | analytik jena | SPEKOL 2000 model | |
| Succinate | Sigma | S2378 | |
| Sucrose | Merck | 1076871000 | |
| Thermomixer | Eppendorph | ||
| Thioflavin T | Sigma | T3516 | |
| Tris-HCl | Merck | 1082191000 | |
| Triton X-100 | Sigma | T9284 | |
| Tryptone | QUELAB | ||
| Water bath | Memmert | ||
| Yeast Extract | QUELAB | ||
| β-NADH | Sigma | N8129 |
References
- Merlini, G., Bellotti, V. Molecular mechanisms of amyloidosis. New England Journal of Medicine. 349, 583-596 (2003).
- Berg, I. . Modeling amyloid disease in Drosophila melanogaster, Linköping Studies in Science and Technology Dissertation No. 1320. , (2010).
- Kagan, B. L., Uversky, V. N., Fink, A. L. Protein aggregation, ion channel formation, and membrane damage. Protein Misfolding, Aggregation, and Conformational Diseases. , 223-236 (2006).
- Demuro, A., et al. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. The Journal of Biological Chemistry. 280, 17294-17300 (2005).
- Kayed, R., et al. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. The Journal of Biological Chemistry. 279, 46363-46366 (2004).
- Manczak, M., Park, B. S., Jung, Y., Reddy, P. H. Differential expression of oxidative phosphorylation genes in patients with Alzheimer's disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Medicine. 5, 147-162 (2004).
- Vila, M., Ramonet, D., Perier, C. Mitochondrial alterations in Parkinson's disease: new clues. Journal of Neurochemistry. 107, 317-328 (2008).
- Petersen, C. A. H., et al. The amyloid β-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proceedings of the National Academy of Sciences of the United States of America. 105, 13145-13150 (2008).
- Devi, L., Raghavendran, V., Prabhu, B. M., Avadhani, N. G., Anandatheerthavarada, H. K. Mitochondrial import and accumulation of α-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. The Journal of Biological Chemistry. 283, 9089-9100 (2008).
- Costa, V., Scorrano, L. Shaping the role of mitochondria in the pathogenesis of Huntington's disease. EMBO Journal. 31, 1853-1864 (2012).
- Vande Velde, C., Miller, T. M., Cashman, N. R., Cleveland, D. W. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 105, 4022-4027 (2008).
- Kagan, B. L., Azimov, R., Azimova, R. Amyloid peptide channels. The Journal of Membrane Biology. 202, 1-10 (2004).
- Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T., Lansbury, P. T. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 418, 291 (2002).
- Sims, N. R., Anderson, M. F. Isolation of mitochondria from rat brain using Percoll density gradient centrifugation. Nature Protocols. 3, 1228-1239 (2008).
- Ghobeh, M., et al. Interaction of Aβ (25-35) Fibrillation Products with Mitochondria: Effect of Small-Molecule Natural Products. Peptide Science. 102, 473-486 (2014).
- Lowry, O. H., Rosebrough, N. J., Farr, A. L., Randall, R. J. Protein measurement with the folin phenol reagent. The Journal of Biological Chemistry. 193, 265-275 (1951).
- Sottocasa, G. L., Kuylenstierna, B., Ernester, L., Bergstrand, A. Separation and some enzymatic properties of the inner and outer membrane of rat liver mitochondria. Methods in Enzymology. 10, 448-463 (1967).
- Hoyer, W., et al. Dependence of a-Synuclein Aggregate Morphology on Solution Conditions. Journal of Molecular Biology. 322, 383-393 (2002).
- Weinreb, P. H., et al. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 35, 13709-13715 (1996).
- Porter, R. R. Partition chromatography of insulin and other proteins. The Biochemical Journal. 53, 320-328 (1953).
- Goldberg, M. E., Rudolph, R., Jaenicke, R. A kinetic study of the competition between renaturation and aggregation during the refolding of denatured reduced egg white lysozyme. Biochemistry. 30, 2790-2797 (1991).
- Young, T. A., Cunningham, C. C., Bailey, S. M. Reactive oxygen species production by the mitochondrial respiratory chain in isolated rat hepatocytes and liver mitochondria: studies using myxothiazol. Archives of Biochemistry and Biophysics. 405, 65-72 (2002).
- Meratan, A. A., Ghasemi, A., Nemat-Gorgani, M. Membrane integrity and amyloid cytotoxicity: a model study involving mitochondria and lysozyme fibrillation products. Journal of Molecular Biology. 409, 826-838 (2011).
- Katebi, B., Mahdavimehr, M., Meratan, A. A., Ghasemi, A., Nemat-Gorgani, M. Protective effects of silibinin on insulin amyloid fibrillation, cytotoxicity and mitochondrial membrane damage. Archives of Biochemistry and Biophysics. 659, 22-32 (2018).
- Fink, A. L. The aggregation and fibrillation of alpha-synuclein. Accounts of Chemical Research. 39, 628-634 (2006).
- Diraviyam, K., Stahelin, R. V., Cho, W., Murray, D. Computer modeling of the membrane interaction of FYVE domains. Journal of Molecular Biology. 328, 721-736 (2003).
- Van Rooijen, B. D., Claessens, M., Subramaniam, V. Lipid bilayer disruption by oligomeric α-synuclein depends on bilayer charge and accessibility of the hydrophobic core. Biochimica et Biophysica Acta. 1788, 1271-1278 (2009).
- Kourie, J. I., Henry, C. L. Ion channel formation and membrane-linked pathologies of misfolded hydrophobic proteins: the role of dangerous unchaperoned molecules. Clinical and Experimental Pharmacology & Physiology. 29, 741-753 (2002).
- Bucciantini, M., et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 416, 507-511 (2002).
- Bolognesi, B., et al. ANS binding reveals common features of cytotoxic amyloid species. ACS Chemical Biology. 5, 735-740 (2010).
- Posse, E., De Arcuri, B. F., Morero, R. D. Lysozyme interactions with phospholipid vesicles: relationships with fusion and release of aqueous content. Biochimica et Biophysica Acta. 1193, 101-106 (1994).
- Roqanian, S., et al. Polyphenols protect mitochondrial membrane against permeabilization induced by HEWL oligomers: possible mechanism of action. International Journal of Biological Macromolecules. 103, 709-720 (2017).
- Ulmer, T. S., Bax, A., Cole, N. B., Nussbaum, R. L. Structure and dynamics of micelle-bound human alphasynuclein. The Journal of Biological Chemistry. 280, 9595-9603 (2005).
- Stockl, M., Fischer, P., Wanker, E., Herrmann, A. Alpha-synuclein selectively binds to anionic phospholipids embedded in liquid-disordered domains. Journal of Molecular Biology. 375, 1394-1404 (2008).
- Devi, L., et al. Mitochondrial import and accumulation of α-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. The Journal of Biological Chemistry. 283, 9089-9100 (2008).
- Ghio, S., Kamp, F., Cauchi, R., Giese, A., Vassallo, N. Interaction of α-synuclein with biomembranes in Parkinson's disease-role of cardiolipin. Progress in Lipid Research. 61, 73-82 (2016).
- Petersen, C. A. H., et al. The amyloid β-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proceedings of National Academy of Sciences of the United States of America. 105, 13145-13150 (2008).
- Costa, V., Scorrano, L. Shaping the role of mitochondria in the pathogenesis of Huntington's disease. EMBO Journal. 31, 1853-1864 (2012).
- Vande Velde, C., Miller, T. M., Cashman, N. R., Cleveland, D. W. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proceedings of National Academy of Sciences of the United States of America. 105, 4022-4027 (2008).
- Oladzad Abbasabadi, A., et al. Disruption of mitochondrial membrane integrity induced by amyloid aggregates arising from variants of SOD1. International Journal of Biological Macromolecules. 61, 212-217 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved