Method Article
Anti-RDL and Anti-mGlutR1 Receptors Antibody Testing in Honeybee Brain Sections using CRISPR-Cas9
* These authors contributed equally
In This Article
Summary
Presented here is a protocol to use the CRISPR-Cas9 system for reducing the production of a protein in the adult honeybee brain to test antibody specificity.
Abstract
Cluster Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) is a gene editing technique widely used in studies of gene function. We use this method in this study to check for the specificity of antibodies developed against the insect GABAA receptor subunit Resistance to Dieldrin (RDL) and a metabotropic glutamate receptor mGlutR1 (mGluRA). The antibodies were generated in rabbits against the conjugated peptides specific to fruit flies (Drosophila melanogaster) as well to honeybees (Apis mellifera). We used these antibodies in honeybee brain sections to study the distribution of the receptors in honeybee brains. The antibodies were affinity purified against the peptide and tested with immunoblotting and the classical method of preadsorption with peptide conjugates to show that the antibodies are specific to the corresponding peptide conjugates against which they were raised. Here we developed the CRISPR-Cas9 technique to test for the reduction of protein targets in the brain 48 h after CRISPR-Cas9 injection with guide RNAs designed for the corresponding receptor. The CRISPR-Cas9 method can also be used in behavioral analyses in the adult bees when one or multiple genes need to be modified.
Introduction
The recently discovered CRISPR/Cas9 system is a powerful tool that has been used to alter genomic DNA in various model systems and organisms. It has accelerated biomedical research and major technological breakthroughs by making genome modification more efficient and robust than previous methods1. Native to S. pyogenes bacteria, the system relies on a Cas9 endonuclease, whose activity leads to double-stranded breaks (DSBs) in DNA, and a guide RNA (gRNA) that directs the Cas9 protein to a specific, sequence-dependent location2. Double-stranded breaks generated by CRISPR/Cas9 can be repaired via non-homologous end-joining (NHEJ), an error-prone process that can lead to frameshifts, or homology direct repair when a donor template is present. The gRNA itself consists of a target-specific CRISPR RNA (crRNA) and a universal trans-activating crRNA (tracrRNA) which can be chemically synthesized and delivered with purified Cas9 nuclease as a ribonucleoprotein complex (RNP)2,3. Fluorescent labeling of the gRNA or Cas9 nuclease can allow for the detection and intracellular visualization of molecular components via fluorescent microscopy4.
In our present work, we take advantage of the CRISPR-Cas9 system to reduce the protein levels in adult honeybee brains. We studied the metabotropic glutamate receptor (mGluR) and anti-mGlutR1 receptor antibodies and the GABAA receptor subunit RDL and anti-RDL antibodies. We developed a simple method to reduce the amount of protein in the brain of the adult honeybee and used it to drive additional tests of the antibodies developed against the corresponding proteins. Monitoring the fluorescence of CRISPR-Cas9 allowed us to estimate the areas and cells involved in the reduction of the protein.
Using this method, we also characterized the anti-mGlutR1 antibodies that were made in rabbits against the conjugated peptide. The honeybee genome encodes a highly conserved AmGluRA (named mGlutR1 according to NCBI nomenclature) metabotropic glutamate receptor5. The honeybee mGlutR1 gene has four predicted splice variants according to the NCBI database. It has been reported that it is expressed in the central nervous system (CNS) of both pupal and adult bee stages and it is involved in long-term memory formation5. Antibodies developed against mGlutR1 can be an essential tool for studying of the glutamatergic system in the learning and memory process in honeybees.
In our studies, we also characterized anti-RDL antibodies developed in rabbits immunized with conjugated peptides from the Apis mellifera RDL receptor subunit. The honeybee Rdl gene, AmRdl (XM_006565102.3, NCBI database), has 14 predicted splice variants. A partially cloned fragment has been reported in the NCBI database AF094822.1. The RDL receptor function and its physiology is well studied in insects6,7,8, including honeybees9,10,11. Antibodies developed against anti-RDL can be an essential tool for studying the GABAergic system in the learning and memory process in honeybees.
An earlier study on the role of octopamine and tyramine receptors used RNAi injected into the brain with a subsequent test of the amount of protein by Western blot12,13. However, RNAi has some significant limitations. There is only a short time window after RNAi injection within which a reduction of protein occurs13. CRISPR-Cas9 was used very recently in honeybee embryos to delete or modify genes in the entire animal14,15,16. We reported the use of CRISPR-Cas9 to reduce the amount of the protein in the adult honeybee. We developed this approach for honeybees because of the ability to couple it to behavioral studies of learning and memory under controlled laboratory conditions17.
In the present work, we developed antibodies against two receptors and tested them on the adult honeybee brain sections after the protein was reduced by CRISPR-Cas9 injection. At the same time, we established an experimental design that allows use of the method for behavioral experiments.
Protocol
The protocol described here follows the animal care guidelines of Arizona State University.
1. Total Protein Isolation from Brains of Apis mellifera
NOTE: Use Apis mellifera New World Carniolan foragers of unknown age for this experiment.
- Place an aluminum mesh screen over the entrance to the hive to capture forager bees17. Capture each bee in a vial with a small hole in each cap. Place the vials containing the bees in ice to lower their body temperature and immobilize them. Leave the bees in ice for no more than 3 min.
- Secure the immobilized bees into previously prepared metal holders. Ensure that the metal holders are constructed so that the bee can be secured with small pieces of duct tape, but still have its back thorax, wings, and head exposed.
CAUTION: Ensure that the bees are fully immobilized before attempting to place them in the holders. - Feed the bees with a 1 M sucrose solution using a 5 mL syringe until they are no longer hungry. Place the secured bees in a box with a wet paper towel to ensure a humid environment.
- Dissect the bee's brain rapidly by cutting off the head with Barraquer Iris scissors (see Table of Materials) and use the scissors to open the head from the front.
- Cut off the brain from the head capsule, take the brain with #5 forceps, and place it in 100 μL of cold (4-8 °C) lysis buffer. The lysis buffer consists of 120 mM Tris-HCl, 2% sodium dodecyl sulfate (SDS), 5% glycerol, 0.2 mM dithiothreitol, 1% Triton X-100, and 1-5 µg/mL of the protease inhibitors PMSF (phenylmethylsulphonylfluoride), aprotinin, benzamidine (pH 6.8) at 4 °C. Homogenize the brain in the lysis solution by turning in a pestle for about 2 min.
- Centrifuge the sample at 12,000 x g for 20 min. Aspirate 90 μL of the supernatant and discard the pellet.
- Take 1 μL of the supernatant to quantitate the total protein using a fluorimeter. The approximate concentration of the total protein was between 2-3 mg/mL per bee. Take 10 μL of the supernatant and add 10 μL of the lysis buffer and 10 μL of a 6x Laemmli buffer18. Spin briefly and boil for 3 min, then cool down on ice. Spin for 1 min at 10,000 x g to remove all debris. One tenth of a bee brain contains approximately 25 ng of total protein.
2. Western Blotting19
- Make 30 mL of 10% running gel containing 12.15 mL of ultrapure distilled water, 7.5 mL of 1.5 M Tris-HCl (pH 8.8), 0.3 mL of 10% SDS, 10 mL of 30% acrylamide-bis acrylamide solution, 0.15 mL of 10% ammonium persulfate (APS), and 20 μL of tetramethylethylenediamine (TEMED).
- Cast the gel between two glass plates separated by spacers.
- Make a 20 mL stacking gel containing 12.1 mL of ultrapure distilled water, 5.0 mL of 0.5 M Tris-HCl (pH 6.8), 0.2 mL of 10% SDS, 2.6 mL of acryl-bis acrylamide, 0.1 mL of 10% APS, and 20 μL of TEMED.
- When the running gel solidifies pour a stacking gel. Carefully add the plastic separator to cast the loading lane, avoiding bubbles. Wait 15-30 min, until the gel is solidified.
- Start loading the gel using 5 μL of protein standards. Load 20 μL of the lysate mixture from step 1.8 per lane, corresponding to ~1/15 of a bee brain or ~16 ng of total protein per lane. Run the samples for 3.5-4 h total at 16 mA in the stacking gel and 32 mA in the running gel. Stop when the dye leaves the gel.
- Transfer the proteins onto nitrocellulose membranes in transfer buffer (25 mM Tris-HCl, 192 mM glycine, 15% methanol) at 0.45 mA for 1 h 30 min at 4 °C.
- To evaluate the efficiency of the protein transfer following SDS-PAGE before immunoblotting, add Ponceau S staining solution (add 0.1 g of Ponceau S and 5 mL of acetic acid to water to a final volume of 100 mL). Store at 4 °C for 1 min and rinse rapidly with distilled water.
- Label each lane with a ballpoint pen, cut the membrane containing two lanes with brain homogenate and one lane with protein marker and place each in a Western blot incubating box. Wash 3x for 5 min each in phosphate buffered saline (PBS) containing 0.1% Tween 20 (PBS-Tw).
- Block the membrane with 10% NGS (1 mL of normal goat serum to 10 mL of PBS-Tw) in a Western blot incubating box for 1 h. Make an anti-mGlutR1 dilution (5 μL of antibody in 10 mL of 10% NGS). Make anti-RDL1 and anti-RDL2 dilutions (5 μL of antibody in 10 mL of 10% NGS each). Replace the blocking solution in each box with the diluted antibodies and leave overnight (16-24 h) at 4 °C.
- Wash the membrane 3x for 5 min each in PBS-Tw. Incubate the membrane with anti-rabbit IgG HRP-conjugated secondary antibodies at 1:10,000 in 10% NGS PBS-Tw for 2 h at room temperature (RT). Wash membranes 3x in PBS-Tw, then 1x in PBS.
- Detect the bands using western chemiluminescent HRP substrate. Mix two substrates 1:1 (v/v) at RT, put all membranes in the same box and cover them with the substrate mix for 2 min (in a dark room with a red light) at RT. Proceed to protein detection using an autoradiography film with several exposure times. Usually one antibody is tested on one membrane containing the same dilution of brain homogenate on two or three lanes and one lane with the weight marker.
3. Immunocytochemical Procedures
- To dissect honeybee brains for immunocytochemistry, immobilize the honeybees in ice for 30 s. After the bees are immobilized, decapitate the bee with scissors and place the head in a solution of 4% paraformaldehyde in PBS. Work under the fume hood.
CAUTION: The body must be carefully disposed of because the abdomen can still sting after decapitation. - Carefully but rapidly remove the antennae, compound eyes, and cut all around the top exoskeleton with Barraquer Iris scissors. Allow the heads to sit in the fixative solution for 10 min. Remove the rest of the exoskeleton of the head and cut all remaining tracheae.
- Place each brain in a 1.5 mL microcentrifuge tube containing at least 1 mL of 4% paraformaldehyde solution and leave overnight at 4-8 °C.
- Make a 7.6% agarose solution by mixing 3.8 g of agarose and 50 mL of distilled water in an Erlenmeyer flask. Microwave the solution until the agarose liquifies.
NOTE: A small piece of tissue paper can be placed in the opening of the flask in order to prevent the agarose solution from overflowing during heating. - Place the fixed honeybee brains (3-4 brains) in a 35 mm Petri dish and remove the excess fixative with tissue paper. Pour the liquid agarose solution over the brains. Orient the brains in the agarose so that the antenna lobes are facing up. Allow the agarose to cool and solidify.
- After the agarose has solidified, cut out blocks of agarose each containing a brain.
- For vibratome sectioning, prepare a 24 well plate with each well containing a basket with a hydrophobic mesh at the bottom. Fill each well with 600 µL of PBS.
- Cut each block into 70 µm cross sections using the vibratome machine and place the sections in the basket containing PBS.
NOTE: Ensure that sections from the same brain are placed in the same basket. - Wash the brain sections 6x for 10 min each with PBS-TX to ensure that no fixative remains in the sections. Place the multiwell plate on an orbital shaker and wash the brains at 210 rpm. Before each wash be sure to replace the PBS-TX solution with fresh PBS-TX solution. Block with 1% normal donkey serum during the last wash.
- To test the anti-mGlutR1 primary antibody, prepare a 1:112 dilution of anti-mGlutR1 antibodies by adding 9 mL of PBS-TX to 80 µL of anti-mGlutR1 antibodies in a 15 mL centrifuge tube. Vortex the tube briefly to mix thoroughly. The working dilution of antibodies was determined in preliminary experiments.
- To test the anti-RDL primary antibody, prepare a 1:100 dilution of anti-RDL antibodies by adding 30 µL of anti-RDL peptide 1, 30 µL of anti-RDL peptide 2, and 6 mL of PBS-TX in a 15 mL centrifuge tube. Vortex the tube briefly to mix thoroughly. The working dilution of antibodies was determined in preliminary experiments.
- Add 800 µL of antibody solution to each well in the plate. Cover the multiwell plate and wrap it in aluminum foil to prevent degradation from light exposure. Place the plate wrapped in aluminum foil on an orbital shaker and shake at 210 rpm for 2 h. Then leave overnight at RT without shaking.
- After the brain sections have been left overnight, wash with PBS-TX for 10 min. Repeat the washing step 6x.
- Prepare the secondary antibodies (anti-rabbit from donkey) by making a 1:225 dilution of secondary antibodies by adding 40 µL of secondary antibodies to 9 mL of PBS-TX.
- Add 800 µL of the secondary antibody dilution to each well. Cover the plate and wrap it in aluminum foil. Place the plate wrapped in aluminum foil on the orbital shaker and shake at 210 rpm for 2 h. Then leave it overnight at RT.
- Wash the brain sections 3x for 10 min each with PBS-TX and 3x with regular PBS solution.
- For embedding the sections in the slides, prepare the mounting media/glycerol embedding solution modified from Rodriguez et al.20. Add 5 g of the mounting medium in 20 mL PBS and stir for 16 h with a magnetic stirrer. Add 10 mL of glycerol and stir for another 16 h with a magnetic stirrer. Centrifuge for 15 min at 4,000 x g, take the liquid homogenous supernatant, and aliquot in 1 mL tubes. Keep at -20 °C
- Embed the sections onto the slides with a drop of the mounting medium prepared in step 3.17, making sure that each slide contains sections from one brain.
- Preadsorption control of immunostaining with conjugated peptides
- For anti-RDL and conjugated peptides, incubate the working dilution of anti-RDL antibodies with the corresponding peptide conjugate overnight at RT on shaker: condition 1) 500 μg peptide conjugated with Keyhole Limpet Hemocyanin (KLH) via glutaraldehyde; condition 2) without any conjugate.
- Centrifuge each mixture for 10 min at 10,000 x g and collect the supernatant from both conditions (step 3.19.1).
- Incubate the serial sections of honeybee brain with each supernatant and process with the secondary antibodies described above. Serial sections are sections that follow each other during the vibratome sectioning procedures, so the same part of the brain will be exposed to positive and negative controls.
- For anti-mGlutR1 and conjugated peptide, incubate the working dilution of anti-mGlutR1 antibodies at RT with the KLH conjugated peptide containing the 10-4 M peptide (condition 1) and without any conjugate (condition 2).
- Centrifuge each mixture for 10 min at 10,000 x g and collect the supernatant from both controls.
- Incubate the serial sections of honeybee brain with supernatant from both conditions and process with the secondary antibodies (steps 3.14-3.15).
- Embed sections from both conditions onto the slide using embedding media (step 3.17).
4. Test of RDL and mGlutR1 Protein Expression by Immunocytochemistry (section 3) in Honeybee Brain After Injection of the Corresponding CRISPR-Cas9 System
- Design the guides using an online CRISPR-Cas9 design tool21 using the genomic DNA sequences of AmRdl (XM_006565102.3) and sequences of mGlutR1 (XM_006566244.3) (Table 1). Order as Cas9 crRNA and Cas-9 tracrRNA with the fluorescent dye ATTO550 at the 5' end. Order the Cas9 Nuclease V3 (See Table of Materials).
- Prepare the components for the CRISPR-Cas 9 system by making a 100 µM stock of each guide crRNA and tracrRNA using nuclease-free water. Aliquot and store at -20 °C. Prepare the working concentration of Cas-9 solution by adding 2.5 µL of Cas 9 nuclease V3 (10 mg/mL) to 47.5 µL of nucleotide free buffer to obtain a final concentration of 0.5 µg/µL. Make gRNA and RNP for injection.
- Prepare gRNA complex formation for each guide RNA separately (guideRNA:tracrRNAATTO550)
- Label a test tube with the name of the gRNA, add 92 µL of nucleotide free buffer, 4 µL of 100 µM CRISPR-Cas9 tracrRNA- 5'ATTO550, and 4 µL of the guide crRNA solution. Mix gently and spin.
NOTE: Example of labeling of gRNA tubes for RDL: GRDL1, GRDL2, GRDL3 (corresponding to RDL guides RNA in Table 1). Example of labeling of gRNA tubes for mGlutR1: GMGL1, GMGL2, GMGL3 (Table 1). - Heat the solution to 95 °C for 5 min to create gRNA. Cool the mixture at RT for 10 min.
- Label a test tube with the name of the gRNA, add 92 µL of nucleotide free buffer, 4 µL of 100 µM CRISPR-Cas9 tracrRNA- 5'ATTO550, and 4 µL of the guide crRNA solution. Mix gently and spin.
- Prepare RNP complex formation (gRNA: S.p Cas9Nuclease), delivery mixtures, and control.
- Label a test tube with the name of the RNP, add 6 µL of gRNA solution and 6 µL of 0.5 µg/µL S.p Cas9 Nuclease V3. Mix gently and incubate the solutions at 37 °C for 10 min before injection.
NOTE: Example of RNP tube labels: RRDL1, RRDL2, RRDL3. - Make a RNPRDLmix. To prepare the final mixture used for injections, add 4 µL of each RNP from RDL together. RNP/RDL mix = 4 µL RRDL1 + 4 µL RRDL + 4 µL RRDL3.
- Make a RNPmGlutR1mix. Mix 4 µL of each RNP from mGlutR1 together to prepare the final mixture used for injections (RNPmGlutR1mix) = 4 µL RMG1 + 4 µL RMG2 + 4 µL RMG3.
NOTE: Example of RNP tube labels: RMG1, RMG2, RMG3. - Make a control noguideRNA solution by mixing 4 µL of tracrRNA, 92 µL of buffer, and 4 µL of water instead of guideRNA (see section 4.3). Mix 6 µL of this noguideRNA solution and 6 µL of 0.5 µg/µL Cas9 Nuclease V3 (step 4.4.1) to produce the control injection solution.
- Label a test tube with the name of the RNP, add 6 µL of gRNA solution and 6 µL of 0.5 µg/µL S.p Cas9 Nuclease V3. Mix gently and incubate the solutions at 37 °C for 10 min before injection.
5. Injection Procedure
- Immobilize each bee as described in step 1.3 and feed them as in step 1.4. Next, prepare two sets of two wooden boxes to release the bees into after injection: a white box (experimental condition) and a black box (control). Each box should contain a small comb and a feeding Petri dish. One side of each box is made of glass to allow for observation.
- Make feeding Petri dishes. Take the cover of a 35 mm Petri dish, place wax inside of the surface, and place the bottom part of the Petri dish on the wax. Secure the plate into the box.
- Using a 5 mL syringe, inject 1M sucrose solution between the cover and the bottom part of the dish. Bees can quickly put their proboscises between the two plates and will stay dry for a 48 h incubation period. Put one dish in each box.
- To prepare the microinjection system, fill the capillaries with mineral oil without any air bubbles. Place the capillaries in the injector holder of the microinjection system. To load the capillary with the desired injection solution, place a hydrophobic film on a flat surface, and pipette the solution onto it. Inject the oil from the capillaries and replace the mixture with the corresponding RNP mix.
- Using the microinjection system, inject 345 nL of RNP mixture solution directly into each bee's median ocelli.
- For RDL-CRISPR-Cas9 injection, inject eight bees with the RNPRDLmix prepared as described in step 4.4.2. Feed them 1M sucrose after the injection. Release them in the white (experimental) box with the small comb and feeding Petri dish for 48 h. Use another eight bees as controls without any injections, feed them, and release them in the black (control) box.
- For mGlutR1 CRISPR-Cas9, inject nine bees with the RNPmGlutR1mix prepared as described in step 4.4.3. For control inject eight bees with RNA mixture without guide (RNAmix_noguide) prepared as described in step 4.4.4. Feed them 1M sucrose after the injection. Release bees from their holders into the white box (experimental condition) or the black box (control) prepared as described in section 5.1.
- Place the boxes in a polystyrene container with wet paper inside for humidity. Leave bees for 48 h and observe 2x per day to ensure that they have enough food and good humidity.
- Dissect the brain of each bee after 48 h (step 3.1) and process for immunocytochemistry as described in section 3. For anti-mGlutR1 immunostainings use step 3.11 and for anti-RDL immunostainings use step 3.12.
- Perform confocal imaging to evaluate the level of fluorescence in the immune-stained brain sections.
- To evaluate the reduction of protein in immunolabeled brains, use confocal image collection at the same level of gain for both control and injected brains.
6. qPCR-based Drop-off Assay to Evaluate the Modified Genomic RDL DNA 48 H After CRISPR Cas9 RNPRDLmix Injection
- Design the primers, control probe, and drop-off probe to evaluate the amount of DNA modified by CRISPR-Cas9 injection. Each primer is designed so that it flanks at least one CRISPR-Cas 9 guide and the amplicon size is 132 bp. The primers and probes used are given below:
RDL1_For: CTCGGAGTGACCACCGT
RDL1_Rev: CAACGAGGCGAACACCAT
RDL1_control_probe: /5HEX/CGA+C+G+TTT+A+C+CT/3IABkFQ/
RDL1_Drop-off_probe: /56-FAM/CCTA+C+G+T+CA+A+GT/3IABkFQ/- Ensure that the primers correspond to unique sequences that are specific to the area. Ensure that the drop-off probe is designed for the area that overlaps with RDL-CRISPR-Cas9 guide.
- To test if there is a modification of the gDNA in the guide area, inject 12 bees in the ocelli with the RNP_RDL mix described in step 5.3.1 and use eight uninjected bees as controls.
- Dissect out the bee brains without the optic lobes 48 h after the injections and extract the gDNA of each injected and control bee brains (without optic lobes) using the kit following the manufacturer's protocol (see Table of Materials).
- Evaluate the quantity, quality, and purity of the extracted gDNA using a spectrophotometer.
- Quantify the relative expression of modified gDNA of AmRDL using the qPCR-based drop-off protocol and real time PCR cycler.
- Resuspend the oligos to 100 µM, dilute the primers to 10 µM, and the probes to 5 µM.
- Set up the PCR reaction in the 96 well plate as shown here for one sample of gDNA (3 repeats): 10 µL of master mix (see Table of Materials), 1 µL of forward primer, 1 µL of reverse primer, 1 µL of control probe, 1 µL of drop-off probe, 2 µL of gDNA (50 ng), 4 µL of nuclease-free water to bring the final reaction volume to 20 µL.
NOTE: Controls are the solution instead of the samples and water instead of the probes. - Set up the cycling program as follows for a real time PCR cycler: 95 °C for 3 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min.
- Evaluate the relative gene modification using the 2-ΔΔCt method, where
 and
and 
7. Relative Quantification of RDL RNA 48 H After RNPRDLmix Injection
- To test if there is a reduction of the RDL mRNA, inject 20 bees in the ocelli with RNP_RDL mix as described in step 5.3.1 and use 12 uninjected bees as the controls. Dissect out the bee brains (without optic lobes) 48 h after the injections, extract the total mRNA from each injected bee, and separate using the manufacturer's protocol (see Table of Materials).
- Remove any DNA residue remaining in the sample using a DNA-free kit (see Table of Materials).
- Evaluate the quality and the purity of the extracted RNA using a spectrophotometer.
- Quantify the expression of AmRDL using a commercially available fluorescent green RT-PCR kit (Table of Materials) on a real time PCR cycler using the manufacturer's protocol for a 384 well plate.
NOTE: In this experiment, previously published primers were used. For AmRDL (AmRDL_F GGTCGATGGGCTACTACCTG; AmRDL_R TCGATCGACTTGACGTAGGA)22 and actin primers as a reference gene [AmActin_F TGCCAACACTGTCCTTTCTG; AmActin_R GAATTGACCCACCAATCCA]23. The relative gene expression was calculated using the 2-ΔΔCt method (step 6.6).
8. qPCR Based drop-off Assay to Evaluate the Modified Genomic DNA 48 H After RNPmGlutR1mix Injection
- Design the primers, control, and drop-off probes to evaluate the amount of DNA modified by CRISPR-Cas9 injection. Design the primers so that they flank at least one CRISPR-Cas 9 guide and the amplicon size is 96 bp.
mGlutR1_For: GGTGAAACGAACGACGGA
AmGlurR1_Rev: GGAGAGAGGGAGCGAGAA
AmGlurR1_control_probe: /5HEX/CGAGG+G+AAA+CGA+GT/3IABkFQ/
AmGlurR1_Drop-off_probe: /56-FAM/CGA+C+A+C+CG+TC/3IABkFQ/
NOTE: Ensure that primers correspond to unique sequences that are specific to the area that should have been modified. The drop-off probe is designed for the area that overlapped with the CRISPR-Cas9 guide. - To test if there is a modification of the gDNA in the guide area, inject 12 bees in the ocelli with RNP_ GlutR1 mix as described in step 5.3.1 and use eight uninjected bees as controls.
- Dissect out the bee brains (without optic lobes) 48 h after the injections and extract the gDNA from each injected and control bee brain (without optic lobes) using the manufacturer's protocol (see Table of Materials).
- Evaluate the quantity, quality, and purity of the extracted gDNA using a spectrophotometer.
- Quantify the relative modification of gDNA AmGlutR1 using qPCR drop-off protocol and for real time PCR cycler as described in step 6.5.
- Evaluate the relative gene modification using the 2-ΔΔCt method, where
 and
and 
9. Quantification of mGlutR1 RNA 48 h After RNPmGlutR1mix Injection
- To test if there is a reduction in the mGlutR1 RNA mRNA, inject six bees in the ocelli with the RNP_RDL mix as described in step 5.3.2 and use six uninjected bees as controls. Dissect out the bee brains (without optic lobes) 48 h after the injections, extract the total mRNA from each injected bee, and separate using the manufacturer's protocol for RNA isolation (see Table of Materials).
- Remove any DNA residue remaining in the sample using a DNA-free kit (see Table of Materials).
- Evaluate the quality and the purity of the extracted RNA using a spectrophotometer.
- Quantify the expression of mGlutR1 using a SYBR Green RT-PCR kit (see Table of Materials) on a real time PCR cycler with the protocol provided for the 384 well kit. Use the following primers for mGlutR1 (mGlut_F CCTCCTCAACGTCTCCTTCATA; mGlut_R TGCCGTTGTGTTCCGATTT) and actin primers as reference gene [AmActin_F TGCCAACACTGTCCTTTCTG; AmActin_R GAATTGACCCACCAATCCA]. Calculate the relative gene expression by using the 2-ΔΔCt method (step 8.6).
Results
Anti-RDL antibody tests
The antibodies were produced against the RDL peptide conjugates as shown in Figure 1A. The first step in characterizing the anti-RDL antibodies is to check the homogenate of the protein extracted from the bee brain using a Western blot with anti-RDL antibodies and a HRP-labeled goat anti-rabbit IgG secondary antibody (Figure 1A, insert). Both anti-RDL antibodies recognized the band located at ~50-60 kD (arrow), corresponding to the estimated weight of the RDL subunit isoform proteins. To demonstrate that anti-RDL antibodies recognized the peptide in the brain slices, we used a preadsorption control (Figure 1B,C). When antibodies were preincubated with conjugated peptides, the staining on the section was absent. This demonstrated that the anti-RDL antibodies recognized the conjugated peptide against which they were raised. In order to demonstrate that the anti-RDL antibodies recognize the protein in the fixed brain tissue, we used CRISPR-Cas9 to knock out the RDL gene that produces the RDL protein in the cells. Figure 1D1-3, shows control frontal bee brain sections that were labeled with anti-RDL antibodies. This bee was not injected with RDL-CRISPR-Cas9 RNP. In Figure 1D1, anti-RDL labels neuropils in the frontal section of the bee brain. The same frontal section in Figure 1D2 shows the absence of fluorescence from ATTO550, because the RDL-CRISPR-Cas9 complex was not injected.
Figure 1E1-E3 shows a brain section from a bee injected with RDL-CRISPR-Cas 9 and then processed with the same amount of antibodies as the control brain in Figure 1D1-D3. The anti-RDL staining was significantly reduced in the whole brain 48 h after the injection, and the distribution of the ATTO550 staining in the brain (Figure 1E2) shows the success of the RDL-CRISPR-Cas 9 injections in the bee median ocelli. The multiple scattered cells of the brain exhibit ATTO550. The successful injection of RDL-CRISPR-Cas 9 reduced the protein expression compared with the control (Figure 1E1-3). From eight immunostained bee brains, only one brain had a high level of distribution of the RDL-CRISPR-Cas9 in the cells of mushroom body, protocerebrum, and antennal lobe, whereas other brains had cell staining with ATTO550 in the mushroom body calyx, central complex, but not antennal lobe. It is important to note that in these bees, reduction anti-RDL immunostaining was not as dramatic as in the brain shown in Figure 1E1.
Next, to estimate the level of the modified RDL gDNA in the bees 48 h after RDL-CRISPR-Cas9 injection, we performed a qPCR drop-off test, where the drop-off probe was designed to match the area of one of the RDL gRNAs. In these experiments, in the bee brains injected with RDL-CRISPR-Cas 9, the relative reduction of the fluorescence corresponded to the number of the modified gDNA in the samples. In our tests the area corresponding to this guide in gDNA in 12 bees injected with RDL-CRISPR-Cas9 were 64 % ± (mean ± 30 %SD) compared with gDNA in noninjected bees (Figure 3A).
Next, to estimate the level of the RDL RNA in the bees 48 h after injection, we performed qRT-PCR in a separate group of bees (Figure 3B). We compared the level of RDL RNA of RDL-CRISPR-Cas 9 injected bees (n = 19) with the level of RNA in bees that were not injected (n = 12). In these experiments, the relative reduction of the mRNA RDL was 59% ± (mean ± 15% SE) compared with the level of RNA in non-injected bees. When we examined the level of RDL RNA in each bee individually, only 13 bees out of 19 bees showed a significant reduction of the RNA. These data indicate that injection of RDL-CRISPR-Cas9 through the ocelli might not always reach a large number of brain cells, which confirms the data with RDL immune-stained RDL-CRISPR-Cas9 injected bees. In these preparations, only one bee out of 8 had RDL CRISP-Cas9 in many brain cells (mushroom body, protocerebrum, and antennal lobe) compared with other bee brains, where the distribution of the RDL-CRISPR-Cas9 was concentrated in cells in the protocerebrum (mushroom body calyx and central complex) but not the antennal lobe (Figure 4A-D).
Anti-mGlutR1 antibodies tests
We used the anti-mGlutR1 antibodies produced in rabbit against conjugated peptides specific to Drosophila melanogaster (Figure 2A). The sequence of this peptide shows a 94% identity with the bee peptide (CLSDKTRFDYFARTVPPD) Figure 2A. First, we checked the antibodies against the bee brain protein using immunoblotting. The bee brain homogenate was separated by 10% SDS-PAGE and electrophoretically transferred from to a nitrocellulose membrane and stained with anti-mGlutR1. The insert in Figure 2A shows two bands with estimated weights (103 and 83 kD) corresponding to two isoforms. When we tested this antibody on honeybee brains, we found that they label neuropilar profiles and cells in the bee brain sections as illustrated in Figure 2B,D. After preadsorption of the anti-mGlutR1 antibody with conjugated-mGlutR1 peptide, the specific staining disappeared in the bee brain slice (Figure 2C). This confirms that anti-mGlutR1 antibodies recognize the peptide (Figure 2C). Next, we injected a mix of mGlutR1-CRISPR-Cas9 in the median ocelli and used the control noguideRNA. In control bees (n = 7), the fluorescence from ATTO550 was not concentrated in the cells. Some brains had scattered fluorescence ATTO550 labeling. Thus, the control preparation in Figure 2D1-3 shows anti-mGlutR1 staining in the brain but not the ATTO550 fluorescence. When mGlutR1-CRISPR-Cas9 was injected into the ocelli and taken up by many cells, the level of fluorescence of the secondary antibodies was significantly reduced in the area that uptakes the functional mGlutR1RNP (Figure 2E1-3). The bees were monitored for 48 h, and one bee from each experimental condition was found dead. Thus, in this experiment, we checked seven control bees and eight CRISPR-Cas9 bees. All the bees injected with CRISPR-Cas9 had cells that took in mGlutR1-CRISPR-Cas9. Most of these cells were in the mushroom body calyx, central complex, and posterior protocerebrum. Only two bees out of seven showed ATTO550 labeling in many cells in the mushroom body, central complex, and the antennal lobe. An example of one of these bees is shown in Figure 2E. The reduction of the level of mGlutR1 staining in these preparations was significant. The other five bees have ATTO550 labeling corresponding to the successful delivery of the mGlutR1 CRISPR-Cas9 in the mushroom body and posterior protocerebrum but not in the antennal lobes.
Next, to estimate the level of the modified mGlutR1 gDNA in the bees 48 h after injection, we performed a qPCR-based drop-off test, where the drop-off probe was designed to be in the area near the mGlutR1 guide. In these experiments, in the bee brains injected with mGlutR1-CRISPR-Cas 9, the relative modification of gDNA in 12 bees were 59% ± (mean ± 33 %SD) compared with gDNA in noninjected bees (Figure 3A).
These results were also confirmed by qRT-PCR tests in a different group of bees, where we estimated the mGlutR1 RNA levels using qRT-PCR in the bees 48 h after injections with RNPmGlutR1mix (Figure 3B). We compared the level of mGlutR1 RNA of the mGlutR1-CRISPR-Cas9 injected bees (n = 6) with the RNA levels in bees that were not injected (n = 6). In these experiments, the relative reduction of the mRNA mGlutR1 in injected bees was 53% ± (mean ± 18% SE) compared with uninjected bees (Figure 3B).
The section from four different bees that expressed the RNP RDL-CRISPR-Cas9 in Kenyon cell of mushroom body is shown in Figure 4A-D. The example bee with ATTO550 fluorescence in the mushroom body and the antennal lobe is shown in Figure 4E,F.
| Guides | Sequences | gRNA | RNP |
| [guideRNA:tracrRNA] | [gRNA:Cas9 Nuclease] | ||
| RDL_Guide1 | ACCGTAACGCGACCCCCGCT | GRDL1 | RRDL1 |
| RDL_Guide2 | AACGTCGATCGACTTGACGT | GRDL2 | RRDL2 |
| RDL_Guide3 | CCATGACGAAACACGTGCCC | GRDL3 | RRDL3 |
| mGlu_Guide 1 | CGAAAGTTATCTGACGGTGT | GMGL1 | RMGL1 |
| mGlu_Guide 2 | TTCAACGAGAGCAAGTTCAT | GMGL2 | RMGL2 |
| mGlu_Guide 3 | GCAAACGTCGGTAGGAGTGA | GMGL3 | RMGL3 |
Table 1: Nucleotide sequences of guides designed for RDL and mGlutR1.
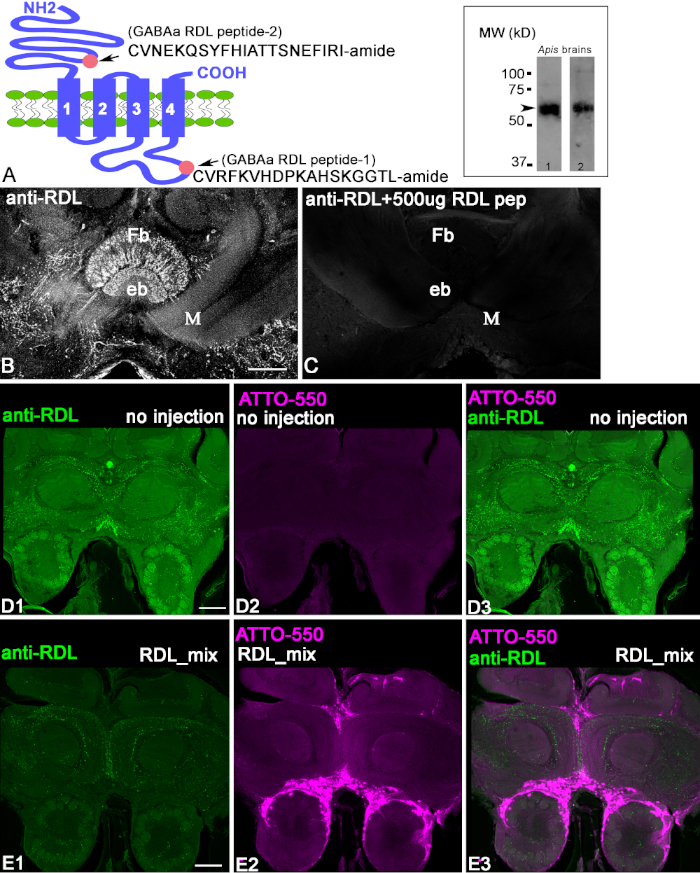
Figure 1: Characterization of anti-RDL antibodies. (A) Schematic of the RDL subunit, where the pink circles indicate the localization of peptide 2 (extracellular CVNEKQSYFHIATTSNEFIRI-amide) in the N-terminus and peptide 1 (intracellular CVRFKVHDPKAHSKGGTL-amide) in the C-terminus. The insert in A shows the bands in the Western blot of honeybee brain extracts processed with corresponding anti-RDL antibodies (one with anti-RDL pep1 and anti-RDL pep2). Each immunoblot shows the apparent size of the protein ~50-60 kD, corresponding to the estimated weights of the various isoform of the RDL subunits. (B,C) Preadsorption of the anti-RDL antibodies with conjugated peptide 1. The image in C shows a reduction in staining in the section when the antibodies were preincubated with conjugated peptide 1. The fan-shaped body (Fb) and Ellipsoid body (eb) are central complex structures in the brain. M = medial lobe of mushroom body. (D1-3) Anti-RDL staining of a control, uninjected bee brain section after 48 h. Green indicates the anti-RDL positive profile in the brain. (D2) This bee was not injected and does not contain ATTO550 fluorescence. (D3) Merged images from D1 and D3. (E1-3) The injection of RDL-CRISPR-Cas9 reduced anti-RDL staining after 48 h. (E2) ATTO550 fluorescence in the cell nuclei indicated successful RDL-CRISPR-Cas9 delivery. (E3) The merged image of anti-RDL (green) and ATTO550 (red). Scale bar = 100 μm (B-E). Please click here to view a larger version of this figure.
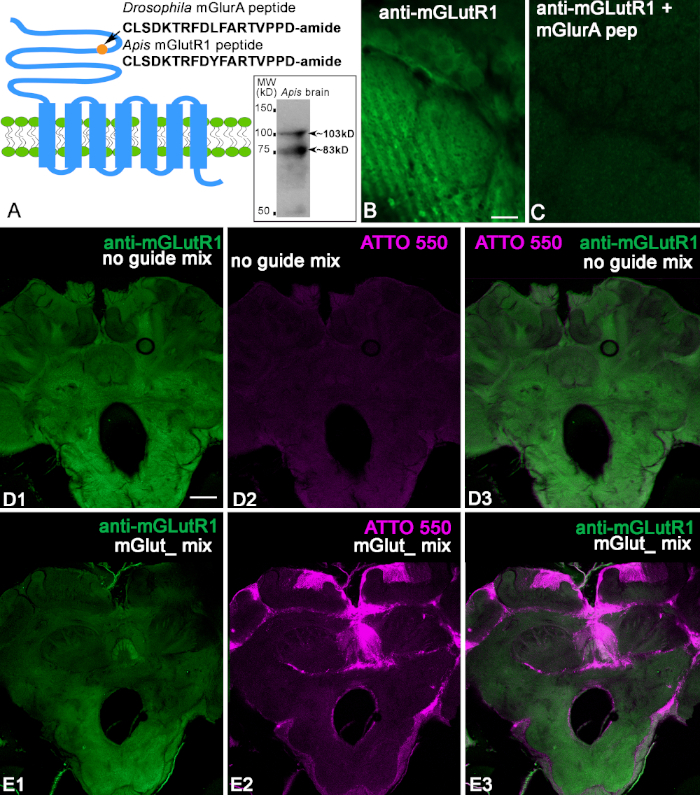
Figure 2: Characterization of anti-mGlutR1 antibodies. (A) Schematic of GCPR mGluR that shows the Drosophila melanogaster peptide used for immunization. For comparison, the Apis mellifera peptide is shown below. The circle indicates the localization of the peptide in the N-terminus of the mGlutR1 receptor extracellular domain. The insert in A shows that the anti-mGlutR1 antibodies recognized two bands in the Western blot of bee brains ~103 kD and ~83 kD that correspond to the estimated weights of known isoforms. (B,C) Preadsorption control of the anti-mGlutR1 antibody in two consecutive sections of the antennal lobe glomeruli. Image of anti-mGlutR1 in the antennal glomeruli section in C shows the reduction of the staining as a result of preincubation of the anti-mGlutR1 peptide with the anti-mGlutR1 antibody. This procedure causes the antibody to precipitate out of solution, which abolishes staining in comparison with B (control, absence of the peptide in the preincubation). (D1) shows staining of anti-mGlutR1 in a bee brain slice after a control injection in the median ocellus. This injection lacked the mGlutR1 gRNA that enables the knock down of mGlutR1 receptors by CRISPR-Cas9, and thus the staining of the anti-mGlutR1 antibody was not reduced (green). (D2) The Absence of ATTO550 fluorescence indicates the absence of functional CRISPR-Cas9 in the brain. (D3) Merged images of anti-mGlutR1 and ATTO550. (E1-E3) show the staining of anti-mGlutR1 in a brain section where mGlutR1 has been permanently knocked down 48 h after injection with mGlutR1-CRISPR-Cas 9 in the median ocellus. Thus, the staining in this brain is greatly reduced due to the successful knockout of the mGlutR1 in many cells. (E2) ATTO550 staining in many cell nuclei in the bee brains indicates that injection of mGlut1-CRISPR-Cas 9 was successful. (E3) Merged image of ATTO550 (red) and anti-mGlutR1 (green). Scale bar = 10 μm (B,C); 100 μm (D,E). Please click here to view a larger version of this figure.
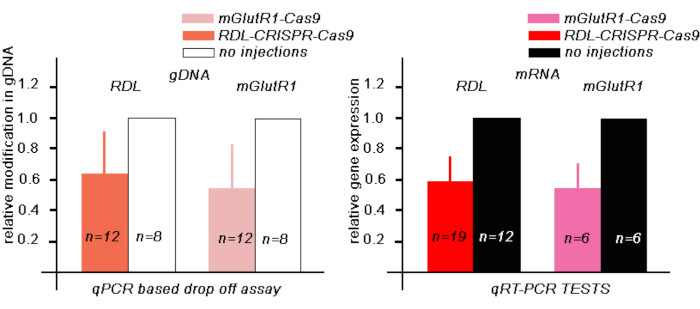
Figure 3: Evaluation of modified gDNA and expression of mRNA of RDL and mGlutR1 mRNA in bee brains 48 h after injection with 345 nL of corresponding RPN CRISPR-Cas9. (A) The qPCR-based drop-off assay test to evaluate the amount of gDNA with a modify area were calculated using 2-ΔΔCt method and normalized against control, uninjected brains. The data are expressed as mean + SD. (B) TheqRT-PCR test was used to evaluate the amount of mRNA in CRISPR-Cas9 injected and uninjected bees. AmActin was used as a reference gene. The relative gene expression was calculated using 2-ΔΔCt methods and normalized against control, uninjected brains. The data are expressed as mean + SE. Please click here to view a larger version of this figure.
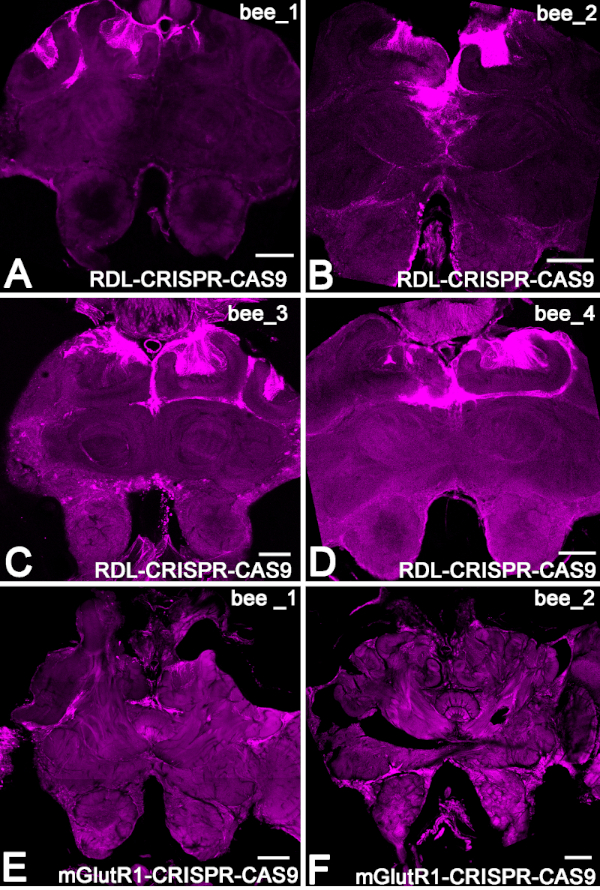
Figure 4: Example of the distribution of RNP CRISPR-Cas9 in bee brains via ATTO550 fluorescence. (A-D) The brain sections from four different bees that expressed the RNP RDL-CRISPR-Cas9 in the Kenyon cell of the mushroom body. (E,F) Example of two brains 48 h after injection with RNPmGlutR1 CRISPR-Cas9. Scale bar = 150 μm (A-F). Please click here to view a larger version of this figure.
Discussion
Characterization of the anti-RDL and anti-mGlutR1
First, we characterized the anti-RDL and anti-mGlutR1 antibodies by immunoblot and pre-adsorption on the slices of fixed honeybee brains. Each antibody was made to recognize all its known isoforms, and Western analysis show they recognize bands that correspond to their predicted molecular weights. Next, both antibodies were blocked by the conjugated peptide against which they were produced on honeybee brain sections.
One of the first aims in our study was to establish that the antibodies produced against the specific conjugated peptide are specific to its protein in fixed brain tissue. For that purpose, we took advantage of the CRISPR-Cas9 system. We designed specific guides for honeybee RDL and mGlutR1 and used each of them to make CRISPR-Cas9 labeled with the fluorescent probe ATTO550. For each receptor, we injected a mixture of three different CRISPR-Cas9 ribonucleoproteins in the ocelli to reduce the amount of the targeted protein in the adult honeybee brain by eliminating the corresponding gene in cells that took up our designed Cas9 system. In our study, we accomplished this step.
One of the first crucial steps for the success of these experiments is designing the appropriate guide RNAs. We recommend designing up to five guide RNAs, located at the beginning, middle, and the end of the gene sequence. In our preliminary work, we tested them in various combinations on three to five bees. We also tried different concentrations of injections, as well as times after injection, and various mixtures of RNP in the injections. We dissected out brains and processed them using anti-RDL and anti-mGlutR1 antibodies. In these initial tests, we established the appropriate combination, post-injection time, as well as the concentration and amount of CRISPR-Cas9 for injection. These initial tests were the basis for setting up the experiments that we described in detail here.
The aim was two-fold: 1) to demonstrate in a bee that our antibody staining was reduced after treatment with CRISPR-Cas9 and 2) to work through the best experimental conditions for behavioral studies. Thus, we show that if many cell nuclei contain CRISPR-Cas9 48 h after injection, the reduction of the anti-RDL and anti-mGlutR1 staining is significant. Additionally, that demonstrates that the tested antibodies specifically recognize the mGlut1 and RDL protein in the honeybee brain preparation and that they can be used for localization studies in the honeybee brain.
Experimental setting CRISPR-Cas9 for behavioral study
Next, we set up the experiments so that CRISPR-Cas9 could be used in behavioral studies. Eight or nine honeybees were collected for control and experimental treatments. They were behaviorally tested before and after injection, and then their brains were processed for ATTO550 and/or immunocytochemistry to determine the brain regions that showed reduction of the target protein. Here it is essential to note that the number of bees taken for one set of experiments was limited to no more than 8-9 bees for the control and experimental conditions. This way both conditions could be tested on the same day. Also, once we prepared the CRISPR-Cas9 mixtures for injection, we never froze them. The CRISPR-Cas9 mixture did not change in potency when used 3 days in a row and kept at 4-8 °C. However, we did not test it after 3 days.
As we described in the Results section for both sets of experimental injections and for both antibodies, only three bees from 16 tested showed a large distribution ATTO550 in the mushroom body, protocerebrum, and antennal lobes. In all other bees, the distribution of CRISPR-Cas9 was limited to the mushroom body, central complex, and/or posterior protocerebrum. It is essential to understand for any behavioral studies that using this injection method the reduction of target protein will be restricted only to the mushroom body in most of the bees. It will not extend to the antennal lobe or subesophageal ganglion. Thus, the injection technique that we use is suitable to study the effect of the reduction of receptors in the mushroom body and central complex in behavioral experiments, while a different method of introducing CRISPR-Cas9 will be more appropriate for studying other brain regions.
In conclusion, our study demonstrated the successful application of CRISPR-Cas9 as a control for antibody staining in the brain. For both antibodies (anti-RDL and anti-mGlutR1), when the uptake of mGlutR1-CRISPR-Cas9 or RDL-CRISPR-Cas9 was successful, the level of corresponding antibody staining was also reduced significantly. Also, it is essential to note that injection in the ocelli led to a distribution of CRISPR-Cas9 in the brain that was not homogenous. The distribution varied from a minimal area surrounding the ocelli and mushroom body to many cells in the whole brain. The variability of the mGlutR1-or RDL-CRISPR-Cas9 uptake by the cells was likely due to variation in the injections. Our data show that the CRISPR-Cas9 system works in honeybees, but the method of injection needs to be improved to reduce the variability of CRISPR-Cas9 uptake across individual bee brains. Within these restrictions, it is now possible to employ this technique to manipulate genes in adult bees for behavioral experiments.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the following awards to BHS: Human Frontiers Science Program; NIH NIGMS (R01 GM113967); NSF Ideas Lab (1556337). The peptide and antibodies for DmGluRA were designed in the laboratory Dr. Serge Birman (Marseille, Luminy, France) when IS was supported by Programme d'Urgence FRM/Postdocs UFP20060306548 from the Fondation pour la Recherche Medicale. We are thankful for Daniela Junqueira Marosi and Alex Hanter from Integrated DNA Technology (IDT) for the help with the design of RDL guides and qPCR drop-off assays.
Materials
| Name | Company | Catalog Number | Comments |
| Acetic Acid | Sigma-Aldrich | A6283_100 mL | western blotting |
| Acrylamide-bis Acrylamide | Bio-Rad | 500 mL 1610156 | western blotting |
| Agarose | Sigma-Aldrich | A0169-250g | used with distilled water to fix honey bee brain in blocks |
| Alexa Fluor 488 AffiniPure F(ab')2 Fragment Donkey Anti-Rabbit IgG (H+L) | Jackson Research Laboratories | 711-546-152 | secondary antibody to reveal the primary antibodies from rabbit |
| Alt-R CRISPR-cas9 tracrRNA-5'ATTO | IDT | 1075934 | with desinged guide RNA, it creates gRNA |
| Alt-R S.p. Cas9 nuclease V3 | IDT | 1081058 | enzyme used to make ribonucleoprotein for CRISPR system |
| Ammonium Persulfate | Bio-RAD | 10 g 1610700 | western blotting |
| Anti-mGlutR1 antibodies | Covalab (FRANCE)/21st Century Biochemical | characterized by authors in present paper | Used for primary incubation of mGlut 1 in honey bees |
| Anti-RDL antibodies | 21st Century Biochemical | characterized by authors in present paper | Used for primary incubation of RDL in honey bees |
| Aprotinin | Sigma-Aldrich | Y0001154 | protease inhibitor |
| Barraquer Iris Scissors 7mm Blade Sharp point | World Precision Instruments | 14128-G | used for dissection honey bee brain |
| Benzamidine | Sigma-Aldrich | 12072 | protease inhibitor |
| Blade (breakable) for blade holder | Fine Science Tool | 10050-00 | dissection for western blotting |
| Blade holder and breaker | Fine Science Tool | 15309 | dissection for western blotting |
| Borosilicate glass capillaries | World Precision Instruments | 1B100 F | for injection procedure |
| Chemiluminescent western blot detection substrate | Bio-Rad | 1705062 | western blotting |
| Chloroform | Sigma-Aldrich | 472476-50mL | RNA isolation |
| Dithiothreitol | Bio-Rad | 1610611 | western blotting |
| DNA easy kit | QIAGEN | 69504 | DNA extractionuj |
| DNA-free kit | INVITROGEN | AM1907 | Used to remove DNA |
| Eppendorf Research plus pipette, 3-pack | Sigma-Aldrich | Z683884 | |
| Falcon 24 Well Polystyrene Multiwell | Falcon | 351147 | Multiwell |
| Flat Bottom Embedding Capsules, Polyethylene | Electron Microscopy Science | 70021 | basket for brain sections |
| Forceps Dumont #5 (pair) | Fine Science Tool | 11254-20 | for dissection of honey bee brain from the head |
| Forceps Dumont #5S (pair) | Fine Science Tool | 11252-00 | to clean up the brain from trachea befor dissection |
| Gene Expression Master Mix | Integrated DNA Technologies | 1055770 | qPCR drop off assay |
| Glutaraldehyde | EMS | 16220 | used for preparation of peptide conjugates for control peabsorption |
| Glycerol | Sigma-Aldrich | G5516-500 mL | western blotting, embedding media |
| Glycine | Bio-Rad | 1610718 | western blotting |
| Hydrophobic filtered nylonmesh | Spectrum Labs | 145910 | for bottom of basket for brain sections |
| Isopropyl alcohol | Sigma-Aldrich | I9030-50mL | RNA isolation |
| Keyhole limpet hemocyanin | Sigma-Aldrich | H7017 | used for preparation of conjugates for control |
| LSM800 cofocal microscope | Zeiss | ||
| mGlu_Guide 1 | IDT | designed guide RNA for CRISPR-Cas 9. Target mGlutR1 genome sequences | |
| mGlu_Guide 2 | IDT | designed guide RNA for CRISPR-Cas 9. Target mGlutR1 genome sequences | |
| mGlu_Guide 3 | IDT | designed guide RNA for CRISPR. Target mGlutR1 genome sequences | |
| methanol | Sigma-Aldrich | 34860 | western blotting |
| 96-well PCR microplate | Applied biosystem | 4346907 | qPCR drop off assay and qRT-PCR |
| 384-well PCR microplate | Sigma-Aldrich | Z374911 | qRT-PCR |
| Mineral oil | Sigma-Aldrich | M5904_500mL | for injection procedure |
| Model p87 Flaming Brown Micropipette Puller | Sutter Instrument Co. | Capillary Preparation | |
| Mowiol 4-88 | Sigma-Aldrich | 81381 | for embedding solution |
| Nanoliter 2000 | World Precision Instruments Nanoliter | Injection Apparatus | |
| Normal Donkey Serum | Jackson Research Laboratories | AB_2337258 | blocking agent for immunocytochemistry |
| Non-immune Goat Serum | Invitrogen | 50-062Z 100 mL | blocking agent for immunoblotting |
| Nuclease-free buffer | IDT | 1072570 | Nuclease-free buffer that is used in the preparation of CRISPR-Cas9 injection |
| Nuclease-free water | IDT | AM9337 | nuclease -free water that is used in preparation of CRISPR-Cas 9 system |
| OrbitalShaker Mp4 | Genemate | ||
| Paraformaldehyde | Sigma-Aldrich | 158127-500 g | used with PBS to make fixative for bee brains |
| Phenylmethylsulphonyl fluoride (PMSF) | Sigma-Aldrich | P7626-250 mg | protease inhibitor |
| Phosphate Buffer Saline | Sigma-Aldrich | P4417-100TAB | Used with Paraformaldehyde as fixative; buffer for antibodies |
References
- Harms, D. W., et al. Mouse Genome Editing Using the CRISPR/Cas System. Current Protocols in Human Genetics. 83 (15), 7-27 (2014).
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816-821 (2012).
- Kouranova, E., et al. CRISPRs for Optimal Targeting: Delivery of CRISPR Components as DNA, RNA, and Protein into Cultured Cells and Single-Cell Embryos. Human Gene Therapy. 27 (6), 464-475 (2016).
- Schubert, M., et al. Fluorescently labeled tracrRNA provides efficient genome editing while allowing cellular microscopy and FACS analysis. Genome Editing. , 1-3 (2017).
- Kucharski, R., Mitri, C., Grau, Y., Maleszka, R. Characterization of a metabotropic glutamate receptor in the honeybee (Apis mellifera): implications for memory formation. Invertebrate Neuroscience. 7 (2), 99-108 (2007).
- Aronstein, K., Auld, V., Ffrench-Constant, R. Distribution of two GABA receptor-like subunits in the Drosophila CNS. Invertebrate Neuroscience. 2 (2), 115-120 (1996).
- Thompson, M., Steichen, J. C., ffrench-Constant, R. H. Conservation of cyclodiene insecticide resistance-associated mutations in insects. Insect Molecular Biology. 2 (3), 149-154 (1993).
- Chung, B. Y., Kilman, V. L., Keath, J. R., Pitman, J. L., Allada, R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Current Biology. 19 (5), 386-390 (2009).
- Taylor-Wells, J., Hawkins, J., Colombo, C., Bermudez, I., Jones, A. K. Cloning and functional expression of intracellular loop variants of the honeybee (Apis mellifera) RDL GABA receptor. Neurotoxicology. 60, 207-213 (2017).
- Jones, A. K., Sattelle, D. B. The cys-loop ligand-gated ion channel superfamily of the honeybee, Apis mellifera. Invertebrate Neuroscience. 6 (3), 123-132 (2006).
- Dupuis, J. P., et al. Homomeric RDL and heteromeric RDL/LCCH3 GABA receptors in the honeybee antennal lobes: two candidates for inhibitory transmission in olfactory processing. Journal of Neurophysiology. 103 (1), 458-468 (2010).
- Farooqui, T., Robinson, K., Vaessin, H., Smith, B. H. Modulation of early olfactory processing by an octopaminergic reinforcement pathway in the honeybee. Journal of Neuroscience. 23 (12), 5370-5380 (2003).
- Guo, X., Wang, Y., Sinakevitch, I., Lei, H., Smith, B. H. Comparison of RNAi knockdown effect of tyramine receptor 1 induced by dsRNA and siRNA in brains of the honeybee, Apis mellifera. Journal of Insect Physiology. 111, 47-52 (2018).
- Roth, A., et al. A genetic switch for worker nutrition-mediated traits in honeybees. PLOS Biology. 17 (3), e3000171 (2019).
- Kohno, H., Suenami, S., Takeuchi, H., Sasaki, T., Kubo, T. Production of Knockout Mutants by CRISPR/Cas9 in the European Honeybee, Apis mellifera L. Zoological Science (BIOONE). 33 (5), 505-512 (2016).
- Hu, X. F., Zhang, B., Liao, C. H., Zeng, Z. J. High-Efficiency CRISPR/Cas9-Mediated Gene Editing in Honeybee Apis mellifera Embryos. G3: Genes, Genomes, Genetics. 9 (5), 1759-1766 (2019).
- Smith, B. H., Burden, C. M. A Proboscis Extension Response Protocol for Investigating Behavioral Plasticity in Insects: Application to Basic, Biomedical, and Agricultural Research. Journal of Visualized Experiments. (91), e51057 (2014).
- Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227 (5259), 680-685 (1970).
- Rodriguez, J., Deinhardt, F. Preparation of a semipermanent mounting medium for fluorescent antibody studies. Virology. 12, 316-317 (1960).
- . Custom Alt-R® CRISPR-Cas9 guide RNA Available from: https://www.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM (2019)
- Bonnafe, E., et al. Effect of a thymol application on olfactory memory and gene expression levels in the brain of the honeybee Apis mellifera. Environmental Science and Pollutant Research (International). 22 (11), 8022-8030 (2015).
- Wang, Y., et al. Regulation of behaviorally associated gene networks in worker honeybee ovaries. Journal of Experimental Biology. 215 (Pt 1), 124-134 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved