A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Isotopic Effect in Double Proton Transfer Process of Porphycene Investigated by Enhanced QM/MM Method
In This Article
Summary
A protocol that uses enhanced QM/MM method to investigate the isotopic effect on the double proton transfer process in porphycene is presented here.
Abstract
The single deuterium substitution in porphycene leads to an asymmetric molecular geometry, which may affect the double proton transfer process in the porphycene molecule. In this study, we applied an enhanced QM/MM method called SITS-QM/MM to investigate hydrogen/deuterium (H/D) isotope effects on the double proton transfer in porphycene. Distance changes in SITS-QM/MM molecular dynamics simulations suggested that the deuterium substituted porphycene adopted the stepwise double proton transfer mechanism. The structural analysis and the free energy shifts of double proton transfer process indicated that the asymmetric isotopic substitution subtly compressed the covalent hydrogen bonds and may alter the original transition state location.
Introduction
The proton transfer process in porphycenes holds potential applications in developing molecular switches, transistors and information storage devices1,2. In particular, tautomerization in porphycenes through double proton transfer process has attracted wide interest in the fields of spectroscopy and photophysics2. The inner hydrogen atoms of porphycene can migrate from one trans isomer to the other equivalent trans isomer through double proton transfer process as shown in Figure 1. Two mechanisms have been proposed for the double proton transfer process: the concerted and the stepwise mechanism3,4. In the concerted double proton transfer process, both proton atoms move to the transition state synchronously in a symmetric way, whereas one proton completes the transfer before the other proton in a stepwise process. Two hydrogen atoms can transfer simultaneously or stepwise depending on the correlation strength between two hydrogen atoms5.
Isotopic substitution has been used to detect the structural properties of molecules and rate constants of reaction kinetics6. Single deuterium substitution in the inner hydrogen of porphycene leads to an asymmetric shape of the molecule. The hydrogen bond may expand or contract because of mass difference between the hydrogen and deuterium atoms. The isotopic substitution introduces a perturbation in the scaffold of porphycene. The question arises that whether asymmetric structure would affect the proton transfer process. Limbach and coworkers reported that the replacement of hydrogen with deuterium will compress both hydrogen bonds, and the cooperative coupling of two hydrogen bonds in porphycene may favor concerted mechanism7, whereas Yoshikawa stated the deuteration would make the stepwise mechanism contribute more than the concerted mechanism8. Experimental techniques, such as force spectroscopy, have been developed to capture tautomerization details in a single porphycene9. However, it is still challenging to determine the atomic details of proton transfer experimentally because of its transient nature.
Theoretical calculations and simulations can act as complementary tools in elucidating the reaction mechanisms of proton transfer. Among different theoretical methods, molecular dynamics (MD) simulations can monitor dynamic motions of each atom, and has been widely used to reveal complex mechanisms in chemical and enzymatic reactions. However, regular MD simulations tend to suffer from insufficient sampling issue, especially when high energy barrier exists in the process of interest. Therefore, enhanced sampling methods have been developed, which include transition path sampling10,11, umbrella sampling (US)12,13, and integrated tempering sampling (ITS)14,15. Combination of different enhanced sampling methods can further increase sampling efficiency16,17,18. To harness the enhanced sampling algorithms in simulating chemical reactions, we have implemented the selective integrated tempering sampling (SITS) method with quantum mechanical and molecular mechanical (QM/MM) potentials recently19. The proposed SITS-QM/MM method combines the advantages from both methods: the SITS method accelerates the sampling and can explore all possible reaction channels without prior knowledge of the reaction mechanism, and QM/MM provides more accurate description of the bond forming and bond breaking process, which cannot be simulated by MM methods solely. The implemented SITS-QM/MM approach has successfully uncovered concerted double proton transfer, uncorrelated and correlated stepwise double proton transfer mechanism in different systems, without pre-defining reaction coordinates19. For porphycene, the stepwise but correlated proton transfer character has been reported19. The hybrid SITS-QM/MM method was used to investigate the isotopic effect in porphycene in our study, and below are the detailed descriptions of the algorithm and protocol of our method.
We have implemented SITS method with hybrid QM/MM potentials. The effective potential of SITS was defined to include the potential energy at different temperatures with the weighting factors nk to cover wider temperature ranges,
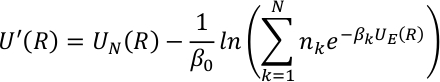
where, N is the number of canonical terms, βk is the inverse temperature, and nk is the corresponding weighting factor for each canonical component. UE(R) and UN(R) represent the enhanced and non-enhanced terms in SITS and are defined as,
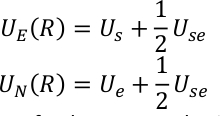
Us, Use and Ue are the potential energy of sub-system, the interaction between sub-system and the environment, and the potential energy of environment. QM/MM potential is expressed as a hybrid summation of three components,
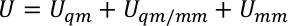
where Uqm, Uqm/mm, and Umm are the internal energy term of the QM subsystem, the interaction energy between the QM and MM regions, and the interaction energy within the MM subsystem, respectively. The Uqm/mm term can be further divided into three components, which include the electrostatic, van der Waals, and covalent interaction energy terms between the QM and MM atoms,

We assign 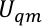 , and
, and  into one Us term in SITS,
into one Us term in SITS,
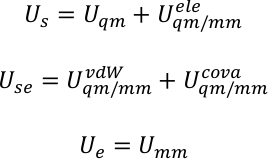
The full potential of the system was then decomposed into the energy of subsystem Us, the interaction energy between the subsystem and the environment Use, and the energy of environment Ue. For instance, in the system of the present work, the subsystem is the porphycence, and the environment the water.
The PMF profile along a collective variable τ(R) is derived as,
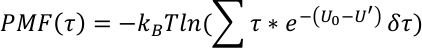
The generally used reaction coordinates for each hydrogen transfer of N1−H1··· N2 are q1 = (r1−r2)/2 and q2 = r1 + r2, where r1 is the distance of N1-H1 and r2 is the distance of H1-N2.
The method has been implemented in the QM/MM MD simulation package QM4D20. The complete source code and documentation can be found here: http://www.qm4d.info/.
Generally, the SITS-QM/MM MD simulations involve four steps: pre-equilibrium (pre-sits); optimization nk (opt-sits); production simulation and data analysis.
Protocol
1. Building model
- Build porphycene structure: Open GaussView software by double-click the mouse. Then click Element Fragment button in the menu of the GaussView to choose the needed elements. Construct porphycene. Then click File button to save as the pdb file.
- Solvate the model: Solvate porphycene in a cubic TIP3P21 water box with an edge length of 38 Å by issuing the command in the linux operating system: genbox_d -cp prp-vac.pdb -cs spc216.gro -o solv.pdb -maxsol 1484 -box 3.8.
- Build deuterium porphycene: Issue the following command to generate topology file: cns < ppi_solv.inp. Then open prp-wat.psf with vi command and change the mass of H1 from 1.00800 to 2.01600 to replace one intramolecular hydrogen atom of porphycene with deuterium to build single deuterium substituted porphycene.
- Set normal MD simulation parameters: Input method scctb, integral 0.5 fs, and cutoff 12 in the MD input file by opening it with vi command.
NOTE: Adopt a cutoff distance of 12 Å for calculating both vdW and electrostatic interactions. Simulate the porphycene molecule with DFTB/MIO method22. Set the integration time step as 0.5 fs for MD simulations. Maintain the temperature of the simulation system at 300 K with Langevin thermostat. Then perform the simulations with QM4D software following the steps below.
2. Pre-sits
- Set up temperature parameters: Input templow 260, temphigh 1100 and ntemp 160 in the input file.
NOTE: The temperature range from 260K to 1100K was spread out by QM4D software to 160 temperature points during MD simulations. The template input files are included in Supplementary Files. - Initiate the pre-sits: Set runtype 100 and step 120,000 in the input file. Then issue the following command: $PATH/qm4d $INPUTFILE > $OUTPUTFILE.
NOTE: The total step is 120,000 but can be adjusted depending on the specific need. The suggested parameters of MD simulations are saved in the $INPUTFILE. The same command is also used in the following opt-sits and production simulation steps, with the input file modified accordingly. - Calculating the decomposed energies
- Extract energy changes: During the pre-sits stage, monitor the energy of each term to calculate the mean values, as shown in Figure 1. Use grep Linux command to extract the energy as follows:
grep ‘SITS-ener0’ $INPUTFILE | awk ‘{a+=$3;b+=$4;c+=$5}END{print a/NR,b/NR,c/NR}. - Modify average energies in the MD input file: Calculate the average energies based on the output of the command line above, and modify the line vshift0 -30801.95; vshift1 -26.88; vshift2 -13888.28 in the input file with the newly generated averages.
NOTE: Numbers -30801.85, -26.88 and -13888.28 are the average energies in the current model system. Please modify the values based on the specific systems.
- Extract energy changes: During the pre-sits stage, monitor the energy of each term to calculate the mean values, as shown in Figure 1. Use grep Linux command to extract the energy as follows:
3. Opt-sits
- Initiate opt-sits: Set runtype 0 in the input file. Then initiate the QM4D program by typing the command as shown in step 2.2 to start the optimization step.
- Monitoring the energy changes and nk values.
- Plot the energy propagation with “grace” program and make sure the energy fluctuation can cover the lowest and the highest ends of the temperature range.
- After optimization, save the final nk values of the opt-sits step into a new file, which is named as nk.dat in this protocol.
4. Running production simulations
- Prepare MD input file: Set runtype 1 in the new input file to start the production simulation step. Specify the file name of stored nk file as nkfile nk.dat in the input file. The number of time steps was set as 6,400,000 in the present systems.
- Initiate production MD simulation: Issue the following command to start MD simulations: $PATH/qm4d $INPUTFILE > $OUTPUTFILE.
NOTE: Make sure the nk values can be read in by the QM4D software. The simulation time is system dependent so change the simulation step based on specific demands. Select a proper number of time steps to ensure enough simulation time for your own system. This step is likely time consuming, so save restart files to avoid restarting the production from the very beginning once being disrupted.
5 Data analysis
- Monitoring the distance changes
- Monitor the bond forming and breaking process during the production phase, use the grep command to check the distance changes of H1-N1 and H1-N2 along the simulation time. The same operation can be conducted for H2-N3 and H2-N4. Then plot the distance propagation using the accumulated distance value during the production simulations.
- Extracting reaction coordinates
- Extract the reaction coordinates and the energy terms from the production output file generated by QM4D by grep command:
grep ‘dist 1’ $OUTPUTFILE | awk ‘{print $5}’ > distance1;
grep ‘ener0’ $OUTPUTFILE > ener0. - Organize data in four columns: q1, q2, U0 and U’ (U0 and U’ are the output normal energy and weighted energy), and write them into the data file at each time frame.
- Extract the reaction coordinates and the energy terms from the production output file generated by QM4D by grep command:
- Calculating Free energy
- Calculate the free energy by issuing the following command:
sits-pmf 300 $INPUTFILE PMF2 [hist_minx hist_maxx num_binsx] [hist_miny hist_maxy num_binsy] > $OUTPUTFILE.
NOTE: sits-pmf is the histogram-based analysis method. [hist_minx hist_maxx num_binsx] defines the range and number of bins for the first reaction coordinate. The second reaction coordinates can be set by [hist_miny hist_maxy num_binsy]. - To project the free energy on the two-dimensional landscape, type the following command:
sits-pmf 300 h1-2d.dat PMF2 -0.6 0.6 24 2.45 4.25 36 > sits-pmf.out.
NOTE: Use a total of 24 bins and 36 bins to cover the distance changes in two selected reaction coordinates, q1 and q2, respectively. Save the 2D PMF data for each hydrogen/deuterium to the sits-pmf.out file.
- Calculate the free energy by issuing the following command:
Results
The single deuterium substitution effect on double proton transfer process in porphycene was examined in the current protocol (Figure 1). The potential energy of QM sub-system and the water during pre-equilibrium and optimization step were checked to make sure the energy has been broadened to a wider energy range (Figure 2). The representative distance and angle changes (Figure 3 and Figure 4), and the ...
Discussion
The structure of porphycene was shown in Figure 1. The electrostatic embedding QM/MM hybrid potential with SITS method was used to describe the chemical reactions in water23,24. The proton transfer occurs within porphycene3 and thus porphycene is set as QM region and the reminding water is set as MM region. Herein we adopted DFTB/MIO as our QM method to treat the porphycene by balancing the efficiency and accu...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research is supported by the National Key Research and Development Program of China (2017YFA0206801, 2018YFA0208600), Natural Science Foundation of Jiangsu Province, and National Natural Science Foundation of China (91645116). L.X is the Zhong-Wu Specially Appointed Professor of the Jiangsu University of Technology. The authors acknowledge the suggestions from Dr. Hao Hu and Dr. Mingjun Yang.
Materials
| Name | Company | Catalog Number | Comments |
| operating system | CentOS Linux release 6.0 | ||
| QM4D software | http://www.qm4d.info/ | in-house program | |
| Computer desktop | HP |
References
- Waluk, J. Ground- and excited-state tautomerism in porphycenes. Accounts of Chemical Research. 39 (12), 945-952 (2006).
- Waluk, J. Spectroscopy and tautomerization studies of porphycenes. Chemical Reviews. 117 (4), 2447-2480 (2017).
- Kozlowski, P. M., Zgierski, M. Z., Baker, J. The inner-hydrogen migration and ground-state structure of porphycene. The Journal of Chemical Physics. 109 (14), 5905-5913 (1998).
- Yoshikawa, T., Sugawara, S., Takayanagi, T., Shiga, M., Tachikawa, M. Theoretical study on the mechanism of double proton transfer in porphycene by path-integral molecular dynamics simulations. Chemical Physics Letters. 496 (1-3), 14-19 (2010).
- Smedarchina, Z., Shibl, M. F., Kühn, O., Fernández-Ramos, A. The tautomerization dynamics of porphycene and its isotopomers - concerted versus stepwise mechanisms. Chemical Physics Letters. 436 (4-6), 314-321 (2007).
- Wolfsberg, M., Hook, W. A., Paneth, P., Rebelo, L. P. N. Isotope effects. The Chemical, Geological, and Bio Sciences. , (2009).
- Pietrzak, M., Shibl, M. F., Bröring, M., Kühn, O., Limbach, H. H. 1H/2H NMR studies of geometric H/D isotope effects on the coupled hydrogen bonds in porphycene derivatives. Journal of the American Chemical Society. 129 (2), 296-304 (2007).
- Yoshikawa, T., Sugawara, S., Takayanagi, T., Shiga, M., Tachikawa, M. Quantum tautomerization in porphycene and its isotopomers: Path-integral molecular dynamics simulations. Chemical Physics. 394 (1), 46-51 (2012).
- Ladenthin, J. N., et al. Force-induced tautomerization in a single molecule. Nature Chemistry. 8 (10), 935-940 (2016).
- Dellago, C., Bolhuis, P. G., Csajka, F. S., Chandler, D. Transition path sampling and the calculation of rate constants. The Journal of Chemical Physics. 108 (5), 1964-1977 (1998).
- Bolhuis, P. G., Chandler, D., Dellago, C., Geissler, P. L. Transition path sampling: Throwing ropes over rough mountain passes, in the dark. Annual Review of Physical Chemistry. 53 (1), 291-318 (2002).
- Torrie, G. M., Valleau, J. P. Nonphysical sampling distributions in monte carlo free-energy estimation: Umbrella sampling. Journal of Computational Physics. 23 (2), 187-199 (1977).
- Kästner, J. Umbrella sampling. Wiley Interdisciplinary Reviews: Computational Molecular Science. 1 (6), 932-942 (2011).
- Gao, Y. Q. An integrate-over-temperature approach for enhanced sampling. Journal of Chemical Physics. 128 (6), 064105 (2008).
- Yang, L., Gao, Y. Q. A selective integrated tempering method. Journal of Chemical Physics. 131 (21), 214109 (2009).
- Yang, M., Yang, L., Gao, Y., Hu, H. Combine umbrella sampling with integrated tempering method for efficient and accurate calculation of free energy changes of complex energy surface. The Journal of Chemical Physics. 141 (4), 044108 (2014).
- Yang, Y. I., Zhang, J., Che, X., Yang, L., Gao, Y. Q. Efficient sampling over rough energy landscapes with high barriers: A combination of metadynamics with integrated tempering sampling. The Journal of Chemical Physics. 144 (9), 094105 (2016).
- Xie, L., Shen, L., Chen, Z. N., Yang, M. Efficient free energy calculations by combining two complementary tempering sampling methods. The Journal of Chemical Physics. 146 (2), 024103 (2017).
- Xie, L., Cheng, H., Fang, D., Chen, Z. N., Yang, M. Enhanced QM/MM sampling for free energy calculation of chemical reactions: A case study of double proton transfer. The Journal of Chemical Physics. 150 (4), 044111 (2019).
- Hu, X., Hu, H., Yang, W. . QM4D: an integrated and versatile quantum mechanical/molecular mechanical simulation package (http://www.qm4d.info/). , (2016).
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., Klein, M. L. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 79 (2), 926-935 (1983).
- Walewski, L., et al. Scc-dftb energy barriers for single and double proton transfer processes in the model molecular systems malonaldehyde and porphycene. International Journal of Quantum Chemistry. 106 (3), 636-640 (2006).
- Bakowies, D., Thiel, W. Hybrid models for combined quantum mechanical and molecular mechanical approaches. The Journal of Physical Chemistry. 100 (25), 10580-10594 (1996).
- Hu, H., Yang, W. Development and application of ab initio QM/MM methods for mechanistic simulation of reactions in solution and in enzymes. Journal of Molecular Structure: THEOCHEM. 898 (1-3), 17-30 (2009).
- Xie, L., Yang, M., Chen, Z. N. Understanding the entropic effect in chorismate mutase reaction catalyzed by isochorismate-pyruvate lyase from pseudomonas aeruginosa (PchB). Catalysis Science, Technology. 9 (4), 957-965 (2019).
- Shibl, M. F., Pietrzak, M., Limbach, H. H., Kühn, O. Geometric H/D isotope effects and cooperativity of the hydrogen bonds in porphycene. ChemPhysChem. 8 (2), 315-321 (2007).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved