A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Induction and Analysis of Oxidative Stress in Sleeping Beauty Transposon-Transfected Human Retinal Pigment Epithelial Cells
In This Article
Summary
We present a protocol for the development and use ofan oxidative stress-model by treating retinal pigment epithelial cells with H2O2, analyzing cell morphology, viability, density, glutathione, and UCP-2 level. It is a useful model to investigate the antioxidant effect of proteins secreted by transposon-transfected cells to treat neuroretinal degeneration.
Abstract
Oxidative stress plays a critical role in several degenerative diseases, including age-related macular degeneration (AMD), a pathology that affects ~30 million patients worldwide. It leads to a decrease in retinal pigment epithelium (RPE)-synthesized neuroprotective factors, e.g., pigment epithelium-derived factor (PEDF) and granulocyte-macrophage colony-stimulating factor (GM-CSF), followed by the loss of RPE cells, and eventually photoreceptor and retinal ganglion cell (RGC) death. We hypothesize that the reconstitution of the neuroprotective and neurogenic retinal environment by the subretinal transplantation of transfected RPE cells overexpressing PEDF and GM-CSF has the potential to prevent retinal degeneration by mitigating the effects of oxidative stress, inhibiting inflammation, and supporting cell survival. Using the Sleeping Beauty transposon system (SB100X) human RPE cells have been transfected with the PEDF and GM-CSF genes and shown stable gene integration, long-term gene expression, and protein secretion using qPCR, western blot, ELISA, and immunofluorescence. To confirm the functionality and the potency of the PEDF and GM-CSF secreted by the transfected RPE cells, we have developed an in vitro assay to quantify the reduction of H2O2-induced oxidative stress on RPE cells in culture. Cell protection was evaluated by analyzing cell morphology, density, intracellular level of glutathione, UCP2 gene expression, and cell viability. Both, transfected RPE cells overexpressing PEDF and/or GM-CSF and cells non-transfected but pretreated with PEDF and/or GM-CSF (commercially available or purified from transfected cells) showed significant antioxidant cell protection compared to non-treated controls. The present H2O2-model is a simple and effective approach to evaluate the antioxidant effect of factors that may be effective to treat AMD or similar neurodegenerative diseases.
Introduction
The model described here, offers a useful approach to evaluate the efficiency ofbiopharmaceutical agents for reducing oxidative stress in cells. We have used the model to investigate the protective effects of PEDF and GM-CSF on the H2O2-mediated oxidative stress on retinal pigment epithelial cells, which are exposed to high levels of O2, and visible light, and the phagocytosis of photoreceptor outer segment membranes, generating significant levels of reactive oxygen species (ROS)1,2. They are considered a major contributor to the pathogenesis of avascular age-related macular degeneration (aAMD)3,4,5,6,7,8. Besides, there is a decrease in RPE-synthesized neuroprotective factors, specifically the pigment epithelium-derived factor (PEDF), insulin-like growth factors (IGFs), and granulocyte macrophage-colony-stimulating factor (GM-CSF) leading to the dysfunction and loss of RPE cells, followed by photoreceptor and retinal ganglion cell (RGC) death3,4,5. AMD is a complex disease that results from the interaction between metabolic, functional, genetic, and environmental factors4. The lack of treatments for aAMD is the major cause of blindness in patients older than 60 years of age in industrialized countries9,10. The reconstitution of the neuroprotective and neurogenic retinal environment by the subretinal transplantation of genetically modified RPE cells overexpressing PEDF and GM-CSF has the potential to prevent retinal degeneration by mitigating the effects of oxidative stress, inhibiting inflammation and supporting cell survival11,12,13,14,15,16. Even though there are several methodologies to deliver genes to cells, we have chosen the non-viral hyperactive Sleeping Beauty transposon system to deliver the PEDF and GM-CSF genes to RPE cells because of its safety profile, the integration of the genes into the host cells' genome, and its propensity to integrate the delivered genes in non-transcriptionally active sites as we have shown previously17,18,19.
Cellular oxidative stress can be induced in cells cultured in vitro by several oxidative agents, including hydrogen peroxide (H2O2), 4-hydroynonenal (HNE), tertbutylhydroperoxide (tBH), high oxygen tensions, and visible light (full spectrum or UV irradiation)20,21. High oxygen tensions and light require special equipment and conditions, which limits transferability to other systems. Agents such as H2O2, HNE, and tBH induce overlapping oxidative stress molecular and cellular changes. We chose H2O2 to test the antioxidant activity of PEDF and GM-CSF because it is convenient and biologically relevant since it is produced by RPE cells as a reactive oxygen intermediate during photoreceptor outer segment phagocytosis22 and it is found in ocular tissues in vivo23. Since the oxidation of glutathione may be partially responsible for the production of H2O2 in the eye, we have analyzed the levels of GSH/glutathione in our studies, which are linked to H2O2-induced oxidative stress and the regenerative capacity of cells21,22. The analysis of glutathione levels is especially relevant since it participates in the anti-oxidative protective mechanisms in the eye24. Exposure to H2O2 is used frequently as a model to examine the oxidative stress susceptibility and antioxidant activity of RPE cells1,25,26,27,28,29,30, and, additionally, it shows similarities to light-induced oxidative stress damage, a "physiological" source of oxidative stress21.
To evaluate the functionality and the effectiveness of neuroprotective factors, we have developed an in vitro model that allows for the analysis to quantify the anti-oxidative effect of growth factors expressed by cells genetically modified to overexpress PEDF and GM-CSF. Here, we show that RPE cells transfected with the genes for PEDF and GM-CSF are more resistant to the harmful effects of H2O2 than are non-transfected control cells, as evidenced by cell morphology, density, viability, intracellular level of glutathione, and expression of UCP2 gene, which codes for the mitochondrial uncoupling protein 2 that has been shown to reduce reactive oxygen species (ROS)31.
Protocol
Procedures for the collection and use of human eyes were approved by the Cantonal Ethical Commission for Research (no. 2016-01726).
1. Cell isolation and culture conditions
- Human ARPE-19 cell line
- Culture 5 x 105 ARPE-19 cells, a human RPE cell line, in Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12 Ham (DMEM/Ham´s F-12) supplemented with 10% fetal bovine serum (FBS), 80 U/mL penicillin, 80 µg/mL streptomycin, and 2.5 µg/mL amphotericin B (complete medium) at 37 °C in a humidified atmosphere of 5% CO2 and 95% air in a T75 flask (for other cell densities see Table 1).
- Change the medium three times per week.
- Once the cells are grown to approximately 90% confluence (evaluated qualitatively), aspirate the medium and wash the cells with sterile 1x PBS.
- Incubate the cells with a 5% Trypsin-2% EDTA solution for 7-10 min at 37 °C (for volumes see Table 1). Monitor detachment visually.
- Stop trypsinization by adding complete medium containing 10% FBS (for volumes see Table 1).
- Transfect the cells (see step 2. of the protocol), sub-cultivate the cells at a ratio of 1:10 (once per week), or seed in a 96-well plate as detailed below (see steps 3.3 and 3.4 of the protocol).
| Medium (mL) | ||||||
| Area (cm²) | Seeding density for ARPE-19 cells (cells/well) | Application | For cell culture | To stop trypsin | Volume of trypsin (mL) | |
| Flask T75 | 75 | 5,00,000 | ARPE-19 cell growth | 10 | 7 | 3 |
| 6 Well plate | 9.6 | 1,00,000 | Seeding of transfected ARPE-19 cells | 3 | 1 | 0.5 |
| 24 Well plate | 2 | 50,000 | Seeding of transfected hRPE cells | 1 | 0.8 | 0.2 |
| 96 Well plate | 0.32 | 5,000 for oxidative stress experiments with transfected cells (Fig. 1) | Oxidative stress experiments | 0.2 | ||
| 3,000 for oxidative stress experiments with non-transfected cells plus proteins (Fig. 1) | ||||||
Table 1: Cell culture volumes. Recommended media volumes for cell culture plates and flasks for the culture of ARPE-19 and primary human RPE cells.
- Primary human RPE cells
- Isolate primary human RPE cells as described by Thumann et al.17, and culture cells in complete medium supplemented with 20% FBS.
- Change the medium twice per week. Once the cells reach confluency (monitored visually), reduce FBS to 1% to avoid overgrowth.
- Transfect the cells (see step 2 of the protocol), or seed in a 96-well plate as detailed below (see steps 3.3 and 3.4 of the protocol).
NOTE: Data presented here was collected from the culture of RPE cells obtained from the eyes of four human donors. Table 2 details the demographics of the donors from the Lions Gift of Sight Eye Bank (Saint Paul, MN). The eyes were enucleated 12.7 ± 5.7 h (mean ± SD) post-mortem after informed consent was obtained in accordance with the Declaration of Helsinki.
| No | age | gender | death to preservation (hours) | death to isolation | cultivation | cultivation | Symbol in graph | |
| (days) | before transfection (days) | after transfection (days) | ||||||
| 2 | 80 | M | 20.7 | 8 | 140 | 36 |  | |
| 3 | 86 | F | 12.8 | 8 | 85 | 45 |  | |
| 4 | 86 | F | 8.5 | 5 | 26 | 133 |  | |
| 8 | 83 | F | 8.9 | 6 | 18 | 27 |  | |
| mean | 83.8 | 12.7 | 6.8 | 67.3 | 60.3 | |||
| SD | 2.9 | 5.7 | 1.5 | 57.0 | 49.1 |
Table 2: Demographics of human donors for retinal pigment epithelial cells.
2. Electroporation of ARPE-19 and primary human RPE cells
- Trypsinize ARPE-19 cells or primary human RPE cells as described in steps 1.1.3-1.1.5 of the protocol.
- Perform electroporation with the commercially available transfection kit (see Table of Materials).
- For transfection of ARPE-19 cells refer to Johnen et al.32 and for primary hRPE to Thumann et al.17. Briefly, resuspend 1 x 105 ARPE-19 cells or 5 x 104 primary hRPE cells in 11 µL of R buffer and add 2 µL of plasmid mixture containing 0.03 µg pSB100X transposase33 and 0.47 µg pT2-CMV-PEDF-His or pT2-CMV-GMCSF-His transposon (ratio transposase:transposon 1:16). For PEDF and GM-CSF double transfected cells, use a ratio of 1:16:16 (0.03 µg pSB100X, 0.47 µg pT2-CMV-PEDF-His, and 0.47 µg pT2-CMV-GMCSF-His). Use the following electroporation parameters: two pulses of 1,350 V for 20 ms (pulse width) for ARPE-19 cells; two pulses of 1,100 V for 20 ms for primary cells.
- Seed 1 x 105 transfected ARPE-19 or 5 x 104 transfected primary hRPE cells in 6-well and 24-well plates, respectively, in medium supplemented with 10% FBS without antibiotics or antimycotics. Add penicillin (80 U/mL), streptomycin (80 µg/mL), and amphotericin B (2.5 µg/mL) with the first medium exchange 3 days after transfection.
- Determine cell growth by weekly microscopical monitoring of the cells. Transfection efficiency is monitored by the analysis of gene expression by RT-PCR, and protein secretion by ELISA and WB (methods explained in Supplementary Material).
NOTE: Transfection efficiency can be evaluated for the first time once the cells reach confluency, i.e., at ~7 days and 4 weeks post-transfection for ARPE-19 cells and primary hRPE cells, respectively. - Seed cells in a 96-well plate as detailed below (see step 3.5 of the protocol).
3. Oxidative stress induction (H2O2 treatment) and neuroprotection (PEDF and/or GM-CSF treatment)
- Preparation of conditioned medium of transfected ARPE-19 cells
- Use ARPE-19 cells transfected with the genes PEDF, GM-CSF, or both (see step 2 of the protocol); culture cells for 28 days as described in step 1.1 of the protocol.
- At 28 days post-transfection, trypsinize cells (see steps 1.1.3-1.1.5 of the protocol), count cells using a Neubauer chamber34,35, and seed 5 x 105 cells in T75 flasks in complete medium as described in step 1.1.1 of the protocol. Exchange the medium when the cell culture is approximately 80% confluent (approximately after 1 week; verified qualitatively). Collect the medium after 24 h.
- Store the medium at -20 °C until use.
NOTE: Sufficient concentration of the recombinant PEDF and GM-CSF in the conditioned medium was verified by WB and quantified by ELISA as described in Supplementary Material.
- Purification of PEDF and GM-CSF from conditioned medium of transfected ARPE-19 cells
- Centrifuge the collected medium from step 3.1.2 at 10,000 x g for 15 min at 4 °C.
- Use the Ni-NTA superflow (see Table of Materials) according to the manufacturer's protocols to purify His-tagged proteins as described below.
- Pipette 30 µL of Ni-NTA mixture into a 1.5 mL tube and centrifuge at 2,600 x g for 30 s and discard the flow-through. Wash the pellet twice with 200 µL of 1x incubation buffer.
- Centrifuge at 2,600 x g for 30 s and discard the flow-through. Add 40 µL of 4x Incubation buffer and resuspend.
- Add 900 µL of centrifuged conditioned medium and incubate at 70 rpm (orbital shaker) for 1 h at RT. Centrifuge at 2,600 x g for 1 min and the discard flow-through.
- Wash the pellet twice with 175 µL of 1x incubation buffer. Centrifuge at 2,600 x g for 30 s and discard the flow-through.
- To elute His-tagged PEDF and GM-CSF proteins, add 20 µL of Elution buffer and incubate at 70 rpm (orbital shaker) for 20 min at RT. Centrifuge at 2,600 x g for 30 s. Keep the supernatant containing recombinant PEDF or GM-CSF.
- Quantify the total protein using the commercially available BCA protein assay kit (see Table of Materials) according to the manufacturer's instructions.
- Store the protein solution at -20 °C until use.
NOTE: Incubation buffer (4x) contains 200 mM NaH2PO4, 1.2 M NaCl, and 40 mM Imidazol; Elution buffer contains 50 mM NaH2PO4, 300 mM NaCl, and 250 mM Imidazol.
- Treatment of non-transfected ARPE-19/primary hRPE cells with conditioned medium plus H2O2 (Figure 1A)
- Seed 3,000 non-transfected ARPE-19 (from step 1.1.6 of the protocol) or primary hRPE (from step 1.2.3 of the protocol) cells per well in 96-well plate and culture in 200 µL of conditioned medium from transfected ARPE-19 cells.
- Culture the cells for 10 days at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. Change the conditioned medium every day. Expose the cells to 350 µM H2O2 for 24 h.
- Evaluate oxidative stress damage and determine the antioxidant effect of PEDF and GM-CSF by quantification of glutathione levels (see step 4.1 of the protocol), microscopy (see step 4.2 of the protocol), and cytotoxicity assay (see step 4.2 of the protocol).
NOTE: The duration of the experiment is 12 days. Clear flat bottom microwell plates are used to evaluate luminescence as well as cell morphology. To simultaneously perform the cytotoxicity and glutathione assay, two plates must be seeded with cells on the same day.
- Treatment of non-transfected ARPE-19/primary hRPE cells with PEDF and GM-CSF growth factors plus H2O2 (Figure 1B)
- Seed 3,000 non-transfected ARPE-19 (from step 1.1.6 of the protocol) or primary hRPE (from step 1.2.3 of the protocol) cells per well (96-well plates with a clear flat bottom) in 200 µL of complete culture medium containing 500 ng/mL recombinant PEDF and/or 50 ng/mL recombinant GM-CSF, purified from the medium of transfected ARPE-19 cells or commercially available. Culture cells for 48 h at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. Renew the medium including PEDF and GM-CSF growth factors daily.
NOTE: Add the growth factors fresh to the medium. - After 48 h of treating the cells with the growth factors, remove the medium and add complete medium containing 350 µM H2O2 plus 500 ng/mL PEDF and/or 50 ng/mL GM-CSF.
- Evaluate oxidative stress damage and determine the antioxidant effect of PEDF and GM-CSF by quantification of glutathione levels (see step 4.1 of the protocol), microscopy (see step 4.2 of the protocol), and cytotoxicity assay (see step 4.2 of the protocol).
NOTE: The duration of the experiment is 3 days.
- Seed 3,000 non-transfected ARPE-19 (from step 1.1.6 of the protocol) or primary hRPE (from step 1.2.3 of the protocol) cells per well (96-well plates with a clear flat bottom) in 200 µL of complete culture medium containing 500 ng/mL recombinant PEDF and/or 50 ng/mL recombinant GM-CSF, purified from the medium of transfected ARPE-19 cells or commercially available. Culture cells for 48 h at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. Renew the medium including PEDF and GM-CSF growth factors daily.
- Treatment of transfected ARPE-19/primary hRPE cells with H2O2 (Figure 1C)
- Verify sufficient gene expression and protein secretion of transfected cells by WB and ELISA as described in the Supplementary Material.
- Remove the medium from the wells containing the transfected cells (see step 2 of the protocol).
- Trypsinize cells as described in steps 1.1.3-1.1.5 of the protocol. Count the cells using a Neubauer chamber34,35.
- Seed 5,000 transfected cells/well in 96-well plate in 200 µL of complete medium. Culture cells for 24 h at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. After 24 h, expose the cells to 350 µM H2O2 for 24 h.
- Evaluate oxidative stress damage and determine the antioxidant effect of PEDF and GM-CSF by quantification of glutathione levels (see step 4.1 of the protocol), microscopy (see step 4.2 of the protocol), cytotoxicity assay (see step 4.2 of the protocol), and determination of UCP2 gene expression (see step 4.3 of the protocol).
NOTE: The duration of the experiment is 2 days.
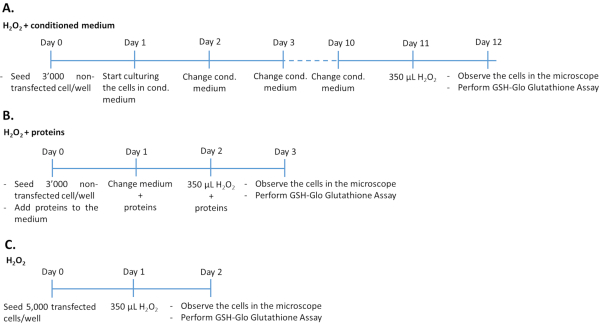
Figure 1: Timelines of the H2O2 assay in the three different experimental approaches. 3,000 non-transfected cells treated with the conditioned medium/recombinant proteins or 5,000 transfected cells were seeded in 96-well plates for treatment with H2O2. To determine the effect of conditioned medium, cells were cultured in 100% cultured medium for 10 consecutive days, changing medium every day. To determine the effect of recombinant growth factors, cells were cultured by adding the appropriate amount of growth factors each day for 3 consecutive days. Note that non-transfected cells were seeded at 3,000 cells per well to avoid overgrowth during the longer culture duration compared to transfected cells. Please click here to view a larger version of this figure.
4. Analysis of oxidative stress level and antioxidant capacity
- Glutathione assay
- Measure the Glutathione (GSH) levels using the commercially available kit (see Table of Materials) following the manufacturer's instructions. Briefly, prepare and appropriate volume of 1x Reagent mix (100 µL reagent/well): Luciferin-NT substrate and Glutathione S-Transferase diluted 1:100 in Reaction Buffer.
NOTE: A 96-well plate requires 10 mL of 1x Reagent mix, which is prepared by adding 100 µL of Luciferin-NT substrate and 100 µL of Glutathione S-Transferase to 10 mL of Reaction buffer. Prepare the 1x Reagent mix immediately before use. Do not store prepared Reagent mix for future use. - Prepare the Luciferin Detection Reagent by transferring one bottle of Reconstitution buffer to the lyophilized Luciferin Detection Reagent.
- Prepare a standard curve using a Glutathione (GSH) standard solution (5 mM). Dilute 5 mM GSH solution 1:100 with dH2O (add 10 µL of 5 mM GSH solution to 990 µL of dH2O). Perform 7 serial 1:1 dilution in 500 µL of dH2O. Transfer 10 µL of each diluted standard to an appropriate well in duplicate.
NOTE: The final concentration of glutathione will range from 0.039 µM to 5 µM. - Prepare the blank (1x Reagent mix) and transfer 10 µL (duplicates) to the appropriate wells.
- Remove the H2O2-treated cells from the incubator.
NOTE: Document the morphology of the H2O2-treated cells by brightfield microscopy (40x).
When the cells are oxidated, they look more rounded and less spread. - Carefully aspirate the culture medium. Add 100 µL of prepared 1x Reagent mix to each well. Mix the cells with the reagent for 15 s at 500 rpm on an orbital shaker.
- Incubate the plate at RT for 30 min. Add 100 µL of reconstituted Luciferin Detection Reagent to each well.
- Mix the solution for 15 s at 500 rpm on an orbital shaker. Incubate the plate for 15 min at RT.
- Determine luminescence using a plate reader using a pre-installed program ADP-Glo.
NOTE: Put the plate inside the plate reader without the lid.- Click on Change Layout and choose the following settings in Basic Parameters: Costar 96-well plate; top optic; positioning delay: 0.1; measurement start time: 0.0; measurement interval time: 1.0; time to normalize the results: 0.0; the gain is adjusted automatically by the device. Define blanks, standards, and samples. Click on Start Measurement.
- Export the data as an Excel file. Calculate the concentration of GSH in each sample by interpolation of the standard curve.
- Measure the Glutathione (GSH) levels using the commercially available kit (see Table of Materials) following the manufacturer's instructions. Briefly, prepare and appropriate volume of 1x Reagent mix (100 µL reagent/well): Luciferin-NT substrate and Glutathione S-Transferase diluted 1:100 in Reaction Buffer.
- Cytotoxicity assay and microscopic analysis
- Aspirate the medium from the cells and add 100 µL of complete medium containing 1% FBS to each well. Return the cells to the incubator.
NOTE: 1% FBS is used because higher percentages of FBS can interfere with the measurement of the luminescence, therefore 1% FBS is used in this case. - Measure cell viability using the commercially available cytotoxicity assay kit (see Table of Materials) following the manufacturer's instructions. Briefly, prepare the Reagent mix adding the Assay buffer to the lyophilized Substrate. Prepare the Lysis Reagent by adding 33 µL Digitonin to 5 mL Assay buffer (for one 96-well plate). Mix well by pipetting up and down to ensure homogeneity.
NOTE: For optimal results, use freshly prepared Reagent mix. Use within 12 h if stored at RT. Reagent mix can be stored at 4 °C for up to 7 days and may be stored in single-use aliquots for up to 4 months at -70 °C. Freezing and thawing must be avoided. The Lysis Reagent can be stored at 4 °C for up to 7 days. - Prepare a standard curve with untreated ARPE-19 cells.
- Trypsinize the cells as described in steps 1.1.3-1.1.5 of the protocol and count the cells using a Neubauer chamber34,35. Centrifuge the cells at 120 g for 10 min at RT. Aspirate the supernatant and resuspend the cell pellet in DMEM/Ham's F12 medium containing 1% FBS to a final concentration of 1 x 105 cells/mL.
- Prepare 7 serial 1:1 dilutions in 200 µL medium containing 1% FBS. Transfer 100 µL of each standard to the appropriate wells (duplicates). Add 50 µL of Reagent mix to all the wells.
- Mix the cells with the reagent for 15 s at 500 rpm on an orbital shaker. Incubate the plate for 15 min at RT. Measure luminescence using the plate reader as described in step 4.1.9 of the protocol. Add 50 µL of the lysis reagent and incubate for 15 min. Measure luminescence using the plate reader as described in step 4.1.9 of the protocol.
- Calculate the percentage of viable cells: (100 - % dead cells) and the percentage of dead cells = [1st luminescence measurement ((dead cells in the sample))/ 2nd luminescence measurement (all cells dead after digitonin treatment)] x 100.
- Aspirate the medium from the cells and add 100 µL of complete medium containing 1% FBS to each well. Return the cells to the incubator.
- UCP2 expression analysis by RT-qPCR
- Trypsinize transfected cells as described above (steps 1.1.3-1.1.5 of the protocol).
- Count the cells using a Neubauer chamber34,35.
- Seed 5,000 transfected ARPE-19 cells/well in 96-well plates.
- After 24 h of culture, treat the cells with 350 µM H2O2 for 24 h.
- Isolate total RNA using a commercial kit for isolation of RNA from low number of cells (see Table of Materials) following the manufacturer's instruction.
- Perform Real-Time quantitative PCR (RT-qPCR) as described in Supplementary Material. Briefly, generate cDNA by retrotranscription using a commercially available mix containing an optimized M-MLV Reverse Transcriptase (see Table of Materials).
- For qPCR employ a ready-to-use reaction cocktail containing all components (including SYBR Green) except primers (see Table S1 of Supplementary Material) and DNA template. Use the following thermocycling conditions: initial denaturation at 95 °C for 10 min, 40 cycles with denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 32 s.
- Use 2^(-ΔΔCT) method for analysis36.
- Preparation of cell lysate for SDS-PAGE and WB analysis of pAkt (Ser473)
- Seed 3 x 105 GM-CSF-transfected ARPE-19 cells/well in 6-well plates (≥21 days post transfection) to determine whether GM-CSF protects RPE cells from damage by H2O2 through the activation of the Akt survival pathway15.
- After 24 h of culture cells are exposed to 350 µM H2O2 for 24 h.
- Mix 1 mL of RIPA buffer with 10 µL of protease phosphatase inhibitor cocktail, 10 µL of 0.5 M EDTA, and 25 µL of 8 M urea (volumes used for one well).
- Carefully aspirate medium and wash the cells with 1x PBS.
- Add the entire volume of RIPA buffer mix to the cells.
- Pipette up and down.
- Collect the lysate in 1.5 mL tubes.
- Centrifuge at 20,000 x g for 30 min at 4 °C.
- Transfer the supernatant to a new 1.5 mL tube.
- Determine the levels of pAkt in 15 µL of undiluted cell lysate by WB as described in Supplementary Material.
Results
Induction of oxidative stress in human Retinal Pigment Epithelial cells
ARPE-19 and primary hRPE cells were treated with varying concentrations of H2O2 for 24 h and the intracellular level of the antioxidant glutathione was quantified (Figure 2A,B). H2O2 at 50 µM and 100 µM did not affect glutathione production, whereas at 350 µM there was a significant decrease...
Discussion
The protocol presented here offers an approach to analyze the anti-oxidative and protective function of PEDF and GM-CSF produced by transfected cells, which can be applied to cells transfected with any putative beneficial gene. In gene therapeutic strategies that have the objective to deliver proteins to tissue by transplanting genetically modified cells, it is critical to obtain information as to the level of protein expression, the longevity of expression, and the effectiveness of the expressed protein in a model of th...
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Gregg Sealy and Alain Conti for excellent technical assistance and Prof. Zsuzsanna Izsvák from the Max-Delbrück Center in Berlin for kindly providing the pSB100X and pT2-CAGGS-Venus plasmids. This work was supported by the Swiss National Sciences Foundation and the European Commission in the context of the Seventh Framework Programme. Z.I was funded by European Research Council, ERC Advanced [ERC-2011-ADG 294742].
Materials
| Name | Company | Catalog Number | Comments |
| 24-well plates | Corning | 353047 | |
| 6-well plates | Greiner | 7657160 | |
| 96-well culture plate white with clear flat bottom | Costar | 3610 | Allows to check the cells before measuring the luminescence (GSH-Glo Assay) |
| 96-well plates | Corning | 353072 | |
| Acrylamid 40% | Biorad | 161-0144 | |
| Amphotericin B | AMIMED | 4-05F00-H | |
| Antibody anti-GMCSF | ThermoFisher Scientific | PA5-24184 | |
| Antibody anti-mouse IgG/IgA/IgM | Agilent | P0260 | |
| Antibody anti-PEDF | Santa Cruz Biotechnology Inc | sc-390172 | |
| Antibody anti-penta-His | Qiagen | 34660 | |
| Antibody anti-phospho-Akt | Cell Signaling Technology | 9271 | |
| Antibody anti-rabbit IgG H&L-HRP | Abcam | ab6721 | |
| Antibody donkey anti-rabbit Alexa Fluor 594 | ThermoFisher Scientific | A11034 | |
| Antibody goat anti-mouse Alexa 488 | ThermoFisher Scientific | A-11029 | |
| ARPE-19 cell line | ATCC | CRL-2302 | |
| BSA | Sigma-Aldrich | A9418-500G | |
| chamber culture glass slides | Corning | 354118 | |
| CytoTox-Glo Cytotoxicity Assay | Promega | G9291 | |
| DAPI | Sigma-Aldrich | D9542-5MG | |
| DMEM/Ham`s F12 | Sigma-Aldrich | D8062 | |
| Duo Set ELISA kit | R&D Systems | DY215-05 | |
| EDTA | ThermoFisher Scientific | 78440 | |
| ELISAquant kit | BioProducts MD | PED613-10-Human | |
| Eyes (human) | Lions Gift of Sight Eye Bank (Saint Paul, MN) | ||
| FBS | Brunschwig | P40-37500 | |
| Fluoromount Aqueous Mounting Medium | Sigma-Aldrich | F4680-25ML | |
| FLUOstar Omega plate reader | BMG Labtech | ||
| GraphPad Prism software (version 8.0) | GraphPad Software, Inc. | ||
| GSH-Glo Glutathione Assay | Promega | V6912 | |
| hydrogen peroxide (H2O2) | Merck | 107209 | |
| ImageJ software (image processing program) | W.S. Rasband, NIH, Bethesda, MD, USA; https://imagej.nih.gov/ij/; 1997–2014 | ||
| Imidazol | Axonlab | A1378.0010 | |
| Leica DMI4000B microscope | Leica Microsystems | ||
| LightCycler 480 Instrument II | Roche Molecular Systems | ||
| LightCycler 480 SW1.5.1 software | Roche Molecular Systems | ||
| NaCl | Sigma-Aldrich | 71376-1000 | |
| NaH2PO4 | Axonlab | 3468.1000 | |
| Neon Transfection System | ThermoFisher Scientific | MPK5000 | |
| Neon Transfection System 10 µL Kit | ThermoFisher Scientific | MPK1096 | |
| Neubauer chamber | Marienfeld-superior | 640010 | |
| Ni-NTA superflow | Qiagen | 30410 | |
| Nitrocellulose | VWR | 732-3197 | |
| Omega Lum G Gel Imaging System | Aplegen Life Science | ||
| PBS 1X | Sigma-Aldrich | D8537 | |
| Penicillin/Streptomycin | Sigma-Aldrich | P0781-100 | |
| PerfeCTa SYBR Green FastMix | Quantabio | 95072-012 | |
| PFA | Sigma-Aldrich | 158127-100G | |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | 23227 | |
| Primers | Invitrogen | See Table 1 in Supplementary Materials | |
| pSB100X (250 ng/µL) | Mátés et al., 2009. Provide by Prof. Zsuzsanna Izsvak | ||
| pT2-CMV-GMCSF-His plasmid DNA (250 ng/µL) | Constructed using the existing pT2-CMV-PEDF-EGFP plasmid reported in Johnen, S. et al. (2012) IOVS, 53 (8), 4787-4796. | ||
| pT2-CMV-PEDF-His plasmid DNA (250 ng/µL) | Constructed using the existing pT2-CMV-PEDF-EGFP plasmid reported in Johnen, S. et al. (2012) IOVS, 53 (8), 4787-4796. | ||
| QIAamp DNA Mini Kit | QIAGEN | 51304 | |
| recombinant hGM-CSF | Peprotech | 100-11 | |
| recombinant hPEDF | BioProductsMD | 004-096 | |
| ReliaPrep RNA Cell Miniprep System | Promega | Z6011 | |
| RIPA buffer | ThermoFisher Scientific | 89901 | |
| RNase-free DNase Set | QIAGEN | 79254 | |
| RNeasy Mini Kit | QIAGEN | 74204 | |
| SDS | Applichem | A2572 | |
| Semi-dry transfer system for WB | Bio-Rad | ||
| SuperMix qScript | Quantabio | 95048-025 | |
| Tris-buffered saline (TBS) | ThermoFisher Scientific | 15504020 | |
| Triton X-100 | AppliChem | A4975 | |
| Trypsin/EDTA | Sigma-Aldrich | T4174 | |
| Tween | AppliChem | A1390 | |
| Urea | ThermoFisher Scientific | 29700 | |
| WesternBright ECL HRP substrate | Advansta | K-12045-D50 | |
| Whatman nitrocellulose membrane | Chemie Brunschwig | MNSC04530301 |
References
- Zareba, M., Raciti, M. W., Henry, M. M., Sarna, T., Burke, J. M. Oxidative stress in ARPE-19 cultures: Do melanosomes confer cytoprotection. Free Radical Biology and Medicine. 40 (1), 87-100 (2006).
- Gong, X., Draper, C. S., Allison, G. S., Marisiddaiah, R., Rubin, L. P. Effects of the macular carotenoid lutein in human retinal pigment epithelial cells. Antioxidants. 6 (4), (2017).
- Sacconi, R., Corbelli, E., Querques, L., Bandello, F., Querques, G. A Review of current and future management of geographic atrophy. Ophthalmology and Therapy. 6, 69-77 (2017).
- Al-Zamil, W. M., Yassin, S. A. Recent developments in age-related macular degeneration: a review. Clinical Interventions in Aging. 12, 1313-1330 (2017).
- Kumar-Singh, R. The role of complement membrane attack complex in dry and wet AMD - From hypothesis to clinical trials. Experimental Eye Research. 184, 266-277 (2019).
- Ung, L., Pattamatta, U., Carnt, N., Wilkinson-Berka, J. L., Liew, G., White, A. J. R. Oxidative stress and reactive oxygen species. Clinical Science. 131, 2865-2883 (2017).
- Beatty, S., Koh, H. H., Phil, M., Henson, D., Boulton, M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Survey of Ophthalmology. 45 (2), 115-134 (2000).
- Alhasani, R. H., et al. Gypenosides protect retinal pigment epithelium cells from oxidative stress. Food and Chemical Toxicology. 112, 76-85 (2018).
- . National Institute of Health Available from: https://nei.nih.gov/learn-about-eye-health/resources-for-health-educators/eye-health-data-and-statistics/age-related-macular-degeneration-amd-data-and-statistics (2020)
- Mitchell, P., Liew, G., Gopinath, B., Wong, T. Y. Age-related macular degeneration. The Lancet. 392, 1147-1159 (2018).
- He, Y., Leung, K. W., Ren, Y., Jinzhi, P., Jian, G., Tombran-Tink, J. PEDF improves mitochondrial function in RPE cells during oxidative stress. Investigative Ophthalmology & Visual Science. 55, 6742-6755 (2014).
- Cao, S., Walker, G. B., Wang, X., Cui, J. Z., Matsubara, J. A. Altered cytokine profiles of human retinal pigment epithelium: Oxidant injury and replicative senescence. Molecular Vision. 19, 718-728 (2013).
- Farnoodian, M., Sorenson, C. M., Sheibani, N. PEDF expression affects the oxidative and inflammatory state of choroidal endothelial cells. Amercian Journal of Physiology and Cell Physiology. 314 (4), 456-472 (2018).
- Polato, F., Becerra, S. P. Retinal Degenerative Diseases: Mechanisms and Experimental Therapies. Retinal Degenerative Diseases. , 699-706 (2016).
- Schallenberg, M., Charalambous, P., Thanos, S. GM-CSF regulates the ERK1/2 pathways and protects injured retinal ganglion cells from induced death. Experimental Eye Research. 89, 665-677 (2009).
- Schallenberg, M., Charalambous, P., Thanos, S. GM-CSF protects rat photoreceptors from death by activating the SRC-dependent signalling and elevating anti-apoptotic factors and neurotrophins. Graefes Archives for Clinical and Experimental Ophthalmology. 250, 699-712 (2012).
- Thumann, G., et al. Engineering of PEDF-expressing primary pigment epithelial cells by the SB transposon system delivered by pFAR4 plasmids. Molecular Therapy - Nucleic Acids. 6, 302-314 (2017).
- Garcia-Garcia, L., et al. Long-term PEDF release in rat iris and retinal epithelial cells after Sleeping Beauty transposon-mediated gene delivery. Molecular Therapy - Nucleic Acids. 9, 1-11 (2017).
- Johnen, S., et al. Antiangiogenic and neurogenic activities of Sleeping Beauty-mediated PEDF-transfected RPE cells in vitro and in vivo. BioMed Research International. 2015, (2015).
- Weigel, A. L., Handa, J. T., Hjelmeland, M. L. Microarray analysis of H2O2-, HNE-, or tBH-treated ARPE-19 cells. Free Radical Biology & Medicine. 33 (10), 1419-1432 (2002).
- Allen, R. G., Tresini, M. Oxidative stress and gene regulation. Free Radical Biology and Medicine. 28 (3), 463-499 (2000).
- Tate, D. J., Miceli, M. V., Newsome, D. A. Phagocytosis and H2O2 induce catalase and metallothionein gene expression in human retinal pigment epithelial cells. Investigative Ophthalmology and Visual Science. 36 (7), 1271-1279 (1995).
- Halliwell, B., Clement, M. V., Long, L. H. Hydrogen peroxide in the human body. FEBS Letters. 486 (1), 10-13 (2000).
- Giblin, F. J., McCready, J. P., Kodama, T., Reddy, V. N. A direct correlation between the levels of ascorbic acid and H2O2 in aqueous humor. Experimental Eye Research. 38, 87-93 (1984).
- Geiger, R. C., Waters, C. M., Kamp, D. W., Glucksberg, M. R. KGF prevents oxygen-mediated damage in ARPE-19 cells. Investigative Ophthalmology and Visual Science. 46, 3435-3442 (2005).
- Campochiaro, P. A., et al. Lentiviral vector gene transfer of endostatin/angiostatin for macular degeneration (GEM) study. Human Gene Therapy. 28, 99-111 (2017).
- Chen, X. -. D., Su, M. -. Y., Chen, T. -. T., Hong, H. -. Y., Han, A. -. D., Li, W. -. S. Oxidative stress affects retinal pigment epithelial cell survival through epidermal growth factor receptor/AKT signaling pathway. International Journal of Ophthalmology. 10 (4), 507-514 (2017).
- Tu, G., et al. Allicin attenuates H2O2 - induced cytotoxicity in retinal pigmented epithelial cells by regulating the levels of reactive oxygen species. Molecular Medicine Reports. 13, 2320-2326 (2016).
- Hao, Y., Liu, J., Wang, Z., Yu, L., Wang, J. Piceatannol protects human retinal pigment epithelial cells against hydrogen peroxide induced oxidative stress and apoptosis through modulating. Nutrients. 11, 1-13 (2019).
- Ballinger, S. W., Van Houten, B., Conklin, C. A., Jin, G. F., Godley, B. F. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Experimental Eye Research. 68 (6), 765-772 (1999).
- Ma, S., et al. Transgenic overexpression of uncoupling protein 2 attenuates salt-induced vascular dysfunction by inhibition of oxidative stress. American Journal of Hypertension. 27 (3), 345-354 (2014).
- Johnen, S., et al. Sleeping Beauty transposon-mediated transfection of retinal and iris pigment epithelial cells. Investigative Ophthalmology and Visual Science. 53 (8), 4787-4796 (2012).
- Mátés, L., et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nature Genetics. 41 (6), 753-761 (2009).
- . Marienfeld Technical information Neubauer-improved Available from: https://www.marienfeld-superior.com/information-about-our-counting-chambers.html (2020)
- . Electron Microscopy Sciences. Neubauer Haemocytometry Available from: https://www.emsdiasum.com/microscopy/technical/datasheet/68052-14.aspx (2020)
- Livak, K. J., Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2^(-ΔΔCT) method. Methods. 25 (4), 402-408 (2001).
- Bascuas, T., et al. Non-virally transfected primary human pigment epithelium cells overexpressing the oxidative stress reduction factors PEDF and GM-CSF to treat retinal neurodegeneration neurodegenerationl. Human Gene Therapy. 30 (11), (2019).
- Zhuge, C. C., et al. Fullerenol protects retinal pigment epithelial cells from oxidative stress-induced premature senescence via activating SIRT1. Investigative Ophthalmology & Visual Science. 55 (7), 4628-4638 (2014).
- Kaczara, P., Sarna, T., Burke, M. Dynamics of H2O2 Availability to ARPE-19 cultures in models of oxidative stress. Free Radical Biology and Medicine. 48 (8), 1068-1070 (2010).
- Gorrini, C., Harris, I. S., Mak, T. W. Modulation of oxidative stress as an anticancer strategy. Nature Reviews Drug Discovery. 12, 931-947 (2013).
- Wang, X., et al. PEDF protects human retinal pigment epithelial cells against oxidative stress via upregulation of UCP2 expression. Molecular Medicine Reports. 19 (1), 59-74 (2019).
- Donadelli, M., Dando, I., Fiorini, C., Palmieri, M. UCP2, a mitochondrial protein regulated at multiple levels. Cellular and Molecular Life Sciences. 71, 1171-1190 (2014).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved