A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Molecular Spring Constant Analysis by Biomembrane Force Probe Spectroscopy
In This Article
Summary
A biomembrane force probe (BFP) is an in situ dynamic force spectroscopy (DFS) technique. BFP can be used to measure the spring constant of molecular interactions on living cells. This protocol presents spring constant analysis for molecular bonds detected by BFP.
Abstract
A biomembrane force probe (BFP) has recently emerged as a native-cell-surface or in situ dynamic force spectroscopy (DFS) nanotool that can measure single-molecular binding kinetics, assess mechanical properties of ligand-receptor interactions, visualize protein dynamic conformational changes and more excitingly elucidate receptor mediated cell mechanosensing mechanisms. More recently, BFP has been used to measure the spring constant of molecular bonds. This protocol describes the step-by-step procedure to perform molecular spring constant DFS analysis. Specifically, two BFP operation modes are discussed, namely the Bead-Cell and Bead-Bead modes. This protocol focuses on deriving spring constants of the molecular bond and cell from DFS raw data.
Introduction
As a live-cell DFS technique, BFP engineers a human red blood cell (RBC; Figure 1) into an ultrasensitive and tunable force transducer with a compatible spring constant range at 0.1-3 pN/nm1,2,3. To probe ligand-receptor interaction, BFP enables DFS measurements at ~1 pN (10-12 N), ~3 nm (10-9 m), and ~0.5 ms (10-3 s) in force, spatial, and temporal resolution4,5. Its experimental configuration consists of two opposing micropipettes, namely the Probe and the target. The Probe micropipette aspirates a RBC and a bead is glued at its apex via a biotin-streptavidin interaction. The bead is coated with the ligand of interest (Figure 1A). The Target micropipette aspirates either a cell or a bead bearing the receptor of interest, corresponding to the Bead-Cell (Figure 1B) and Bead-Bead (Figure 1C) modes, respectively5.
BFP construction, assembly and the DFS experimental protocols were described in detail previously1,6. Briefly, a BFP touch cycle consists of 5 stages: Approach, Impinge, Contact, Retract and Dissociate (Figure 1D). The horizontal RBC apex position is denoted as ΔxRBC. At the beginning, the unstressed (zero-force) RBC deformation ΔxRBC is 0 (Table 1). The Target is then driven by a piezotranslator to impinge on and retract from the Probe bead (Figure 1D). The RBC probe is first compressed by the Target with negative RBC deformation ΔxRBC < 0. In a Bond event, the Retract stage transitions from a compressive to a tensile phase with positive RBC deformation ΔxRBC > 0 (Figure 2C and D). According to Hooke's law, the BFP bearing force is able to be measured as F = kRBC × ΔxRBC, where kRBC (Table 1) is the RBC spring constant of the BFP. Upon bond rupture and the completion of one touch cycle, the probe bead returns to zero-force position with ΔxRBC = 0 (Figure 1D).
To determine the kRBC, we measure and record the radii of the probe micropipette inner orifice (Rp), the RBC (R0) and the circular contact area (Rc) between the RBC and the probe bead (Figure 1A). Then kRBC is calculated according to the Evan's model (Eq. 1)7,8 using a LabVIEW program that acts as a virtual instrument (VI) to operate the BFP (Figure S1A)8,9.
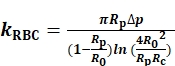 (Eq. 1)
(Eq. 1)
With a BFP established and DFS raw data obtained, hereby we present how to analyze the spring constant of ligand-receptor pair or cells. The DFS raw data on the interaction of the glycosylated protein Thy-1 and K562 cell bearing integrin α5β1 (Thy-1-α5β1; Figures 3A and 3B)10 and that of the fibrinogen and bead coated integrin αIIbβ3 (FGN-αIIbβ3; Figure 3C)11,12 have been used to demonstrate the Bead-Cell and Bead-Bead analysis modes, respectively.
BFP Experimental Preparation
For details of BFP experimental preparation and instrumentation, please refer to the previously published protocols3. In brief, human RBC has been biotinylated using the Biotin-PEG3500-NHS in the carbon/bicarbonate buffer. Proteins of interest have been covalently coupled to the borosilicate glass beads using MAL-PEG3500-NHS in the phosphate buffer. To attach to the biotinylated RBC, the probe bead is also coated with streptavidin (SA) using the MAL-SA. Please see the Table of Materials and Table 2.
To assemble the BFP (Figure 1, left), the third micropipette termed 'Helper' will be used to deliver the probe bead and glue it to the RBC's apex1,3. The covalent interaction between the SA coated probe bead and biotinylated RBC is much stronger than the ligand-receptor bond of interest. Thus, the Dissociate stage can be interpreted as the ligand-receptor bond rupture rather than the detachment of Probe bead from the RBC.
Protocol
1. Obtain Analyzable DFS Events
- Start the experiment in the software (e.g., LabVIEW VI) for the BFP control and parameter setting (Figure S1A).
- Observe the repetitive probe bead-target bead/cell touches in the software for BFP Monitor (Figure S1B).
- Test and achieve the adhesion frequency ≤ 20% within the first 50 touches by tuning the impingement force and contact time, by which it ensures that ≥ 89% of DFS adhesion event are mediated by single bonds12,13,14.
NOTE: For each Bead-Cell/Bead pair, we perform 200 repetitive touch cycles. To obtain publishable data quality, we usually perform n ≥ 3 Bead-Bead or Bead-Cell pairs.- Save data, in the form of Force vs. Time, to the user directed folder by the end of each pair, prompted by the software for BFP control and parameter setting.
- Collect the Force vs. Time raw data of Bond events, as exemplified in the Figure 2A, using the BFP acquisition platform (Figure S1C).
- Open the BFP data analysis software. Click on the yellow folder icon and select the corresponding raw data file by double clicking on them.
- Run the program, and then click the up and down button to switch between events. Use the outlier exclusion criteria (Figure S2) to screen out invalid events. Select the exporting data type as Force vs. Time format and click on the Export Plots Data button.
2. Convert the Force vs. Time Curve to the Force vs. Displacement Curve
- Export the data segment corresponding to the Retract stage to a spreadsheet (Figure 2A, square marquee), which is relevant to the spring constant analysis.
- Plot the Force vs. Time Curve using spreadsheet software. To obtain the Force vs. Displacment curve, convert the time values (Figure 2A, x-axis) to the total displacement values (Δxtot) by multiplying time values with piezo movement velocity (i.e., 4,000 nm/s by preset).
- Zero the first data point by subtracting the smallest displacement value from each acquired displacement value. This horizontal transformation does not affect the ascending slopes of the Retract stage nor the subsequent spring constant calculation.
- Notably, the BFP is considered as a serial spring system in which Δxtot (Table 1) sum deformations of the RBC, ΔxRBC (Table 1), the molecular bond, Δxmol (Table 1), and the Target cell, Δxcell (Table 1), as the Eq. 2:
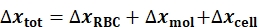 (Eq. 2)
(Eq. 2) - Plot the Force (F) vs. Displacement (Δxtot) curve as shown in the Figure 2B.
3. Spring Cnstant Analysis of Bead- Cell Mode
- In the Force vs. Displacement curve, two distinct slop can be identified, where each can represent the compressive phase and the tensile phase. Fit a regression line to each data group (Figure 2B), where the larger linear fit slope represents the total spring constant at compressive phase (Figure 2B, red), denoted as k1 (Table 1); and the smaller linear fit slope represents the total spring constant at tenslile phase (Figure 2B, blue), denotated as k2 (Table 1).
- For springs connected in series per step 2.2 description, express the reciprocal of the total spring constant, ktot (Table 1), as the sum of the spring constant inverses of RBC, kRBC (Table 1), the molecular bond, kmol (Table 1), and the Target cell, kcell (Table 1). During the compressive phase of the Bead-Cell mode, the molecular bond is not stretched, therefore kmol is not taken into consideration. The reciprocal of the ktot in this scenario (1/k1) is expressed as
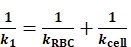 (Eq. 3).
(Eq. 3).
In the example data, kRBC is pre-determined (0.25 pN/nm by default). kcell can be derived from the Eq. 3 with the acquired k1 and kRBC (Figure 3B). - During the tensile phase, adhesion is formed between the ligand-receptor pair. Express the reciprocal of the ktot in this scenario (1/k2) as
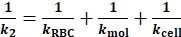 (Eq. 4)
(Eq. 4)
where k2 (Table 1) represents the total spring constant during the tensile phase. - Derive kmol from subtracting 1/k1 from 1/k2 (compare Eq. 3 vs. Eq. 4).
4. Spring Constant Analysis of Bead- Bead Mode
- Fit a regression line to the compressive phase data to obtain k1 (similar to the Figure 2B, red). Of note, in the Bead-Bead mode, the Target cell is replaced by a glass bead coated with the receptor of interest (Figure 1C). Since bead deformation is negligible, the 1/kcell term can be removed from the Eq. 3 and Eq. 4 accordingly. The reciprocal ktot of the compressive phase (1/k1) can be expressed as:
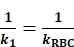 (Eq. 5)
(Eq. 5) - Fit a regression line to the tensile phase data to obtain k2 (similar to the Figure 2B, blue). The reciprocal ktot of the tensile phase (1/k2) can be expressed as:
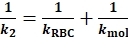 (Eq. 6)
(Eq. 6) - Derive kmol from subtracting 1/k1 from 1/k2 (compare Eq. 5 vs. Eq. 6).
Results
In this work, we have demonstrated the protocol of the BFP spring constant analysis. For the Bead-Cell analysis mode, we analyzed the kmol of the molecular bond between the glycosylated protein Thy-1 coated on the Probe bead and the integrin α5β1 expressed on the Target K562 cell (Thy-1-integrin α5β1; Figure 3A)10. The kcell is also derived from the Bead-Cell mode (K562 Cell;
Discussion
In summary, we have provided a detailed data analysis protocol for preprocessing the DFS raw data and deriving molecular spring constants in the BFP Bead-Bead and Bead-Cell analysis modes. Biomechanical models and equations required for determining molecular and cellular spring constants are presented. Albeit different integrins are studied, the kmol measured by the Bead-Bead mode and the Bead-Cell mode possesses significant range differences (Figure 3A vs.
Disclosures
The authors declare that they have no competing interests to report regarding the present study.
Acknowledgements
We thank Guillaume Troadec for helpful discussion, Zihao Wang for hardware consultation, and Sydney Manufacturing Hub, Gregg Suaning and Simon Ringer for support of our lab startup. This work was supported by Australian Research Council Discovery Project (DP200101970 - L.A.J.), NSW Cardiovascular Capacity Building Program (Early-Mid Career Researcher Grant - L.A.J.), Sydney Research Accelerator prize (SOAR - L.A.J.), Ramaciotti Foundations Health Investment Grant (2020HIG76 - L.A.J.), National Health and Medical Research Council Ideas Grant (APP2003904 - L.A.J.), and The University of Sydney Faculty of Engineering Startup Fund and Major Equipment Scheme (L.A.J.). Lining Arnold Ju is an Australian Research Council DECRA fellow (DE190100609).
Materials
| Name | Company | Catalog Number | Comments |
| 3-Mercaptopropyltrimethoxysilane (MPTMS) | Uct, Specialties, llc | 4420-74-0 | Glass bead functionalization |
| Anhy. Sodium Phosphate Dibasic (Na2HPO4) | Sigma-Aldrich | S7907 | Phosphate buffer preparation |
| BFP data acquisition VI | LabVIEW | BFP control and parameter setting | |
| BFP data analysis VI | LabVIEW | BFP raw data analysis | |
| Biotin-PEG3500-NHS | JenKem | A5026-1 | RBC biotinylation |
| Borosilicate Glass beads | Distrilab Particle Technology, Netherlands | 9002 | Glass bead functionalization |
| Bovine serum albumin | Sigma-Aldrich | A0336 | Ligand functionalization |
| Camera VI | LabVIEW | BFP monitoring | |
| D-glucose | Sigma-Aldrich | G7021 | Tyrode’s buffer preparation |
| Hepes | Sigma-Aldrich | H3375 | Tyrode’s buffer preparation |
| MAL-PEG3500-NHS | JenKem | A5002-1 | Glass bead functionalization |
| Potassium Chloride (KCl) | Sigma-Aldrich | P9541 | Tyrode’s buffer preparation |
| Sodium Bicarbonate (NaHCO3) | Sigma-Aldrich | S5761 | Carbonate/bicarbonate buffer preparation; Tyrode’s buffer preparation |
| Sodium Carbonate (Na2CO3) | Sigma-Aldrich | S2127 | Carbonate/bicarbonate buffer preparation |
| Sodium Chloride (NaCl) | Sigma-Aldrich | S7653 | Tyrode’s buffer preparation |
| Sodium Phosphate Monobasic Monohydrate (NaH2PO4•H2O) | Sigma-Aldrich | S9638 | Phosphate buffer preparation |
| Streptavidin-Maleimide | Sigma-Aldrich | S9415 | Glass bead functionalization |
References
- Chen, Y., et al. Fluorescence Biomembrane Force Probe: Concurrent Quantitation of Receptor-ligand Kinetics and Binding-induced Intracellular Signaling on a Single Cell. The Journal of Visualized Experiments. (102), e52975 (2015).
- Su, Q. P., Ju, L. A. Biophysical nanotools for single-molecule dynamics. Biophysics Reviews. 10 (5), 1349-1357 (2018).
- Ju, L. Dynamic Force Spectroscopy Analysis on the Redox States of Protein Disulphide Bonds. Methods in Molecular Biology. 1967, 115-131 (2019).
- An, C., et al. Ultra-stable Biomembrane Force Probe for Accurately Determining Slow Dissociation Kinetics of PD-1 Blockade Antibodies on Single Living Cells. Nano Letters. 20 (7), 5133-5140 (2020).
- Chen, Y., Ju, L., Rushdi, M., Ge, C., Zhu, C. Receptor-mediated cell mechanosensing. Molecular Biology of the Cell. 28 (23), 3134-3155 (2017).
- Ju, L., Chen, Y., Rushdi, M. N., Chen, W., Zhu, C. Two-Dimensional Analysis of Cross-Junctional Molecular Interaction by Force Probes. Methods in Molecular Biology. 1584, 231-258 (2017).
- Evans, E., Ritchie, K., Merkel, R. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophysical Journal. 68 (6), 2580-2587 (1995).
- Ju, L., Zhu, C. Benchmarks of Biomembrane Force Probe Spring Constant Models. Biophysical Journal. 113 (12), 2842-2845 (2017).
- Evans, E., Ritchie, K., Merkel, R. Sensitive Force Technique to Probe Molecular Adhesion and Structural Linkages at Biological Interfaces. Biophysical Journal. 68, 2580 (1995).
- Fiore, V. F., Ju, L., Chen, Y., Zhu, C., Barker, T. H. Dynamic catch of a Thy-1-alpha5beta1+syndecan-4 trimolecular complex. Nature Communications. 5, 4886 (2014).
- Passam, F., et al. Mechano-redox control of integrin de-adhesion. Elife. 7, (2018).
- Chen, Y., et al. An integrin alphaIIbbeta3 intermediate affinity state mediates biomechanical platelet aggregation. Nature Materials. 18 (7), 760-769 (2019).
- Chen, Y., Lee, H., Tong, H., Schwartz, M., Zhu, C. Force regulated conformational change of integrin αVβ3. Matrix Biology. 60, 70-85 (2017).
- Liu, B., Chen, W., Zhu, C. Molecular force spectroscopy on cells. Annual Review of Physical Chemistry. 66, 427-451 (2015).
- Piper, J. W., Swerlick, R. A., Zhu, C. Determining force dependence of two-dimensional receptor-ligand binding affinity by centrifugation. Biophysical Journal. 74 (1), 492-513 (1998).
- Ju, L., Dong, J. -. f., Cruz, M. A., Zhu, C. The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ibα. Journal of Biological Chemistry. 288 (45), 32289-32301 (2013).
- Ju, L., Chen, Y., Xue, L., Du, X., Zhu, C. Cooperative unfolding of distinctive mechanoreceptor domains transduces force into signals. Elife. 5, 15447 (2016).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved