A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Measuring the Time-Evolution of Nanoscale Materials with Stopped-Flow and Small-Angle Neutron Scattering
In This Article
Summary
This protocol presents the use of a stopped-flow sample environment to quickly mix multiple liquid solutions in situ during a small-angle neutron scattering measurement and to study kinetic processes on nanometer length scales and second time scales.
Abstract
This paper presents the use of a stopped-flow small-angle neutron-scattering (SANS) sample environment to quickly mix liquid samples and study nanoscale kinetic processes on time scales of seconds to minutes. The stopped-flow sample environment uses commercially available syringe pumps to mix the desired volumes of liquid samples that are then injected through a dynamic mixer into a quartz glass cell in approximately 1 s. Time-resolved SANS data acquisition is synced with the sample mixing to follow the evolution of the nanostructure in solution after mixing.
To make the most efficient use of neutron beam time, we use a series of flow selector valves to automatically load, rinse, and dry the cell between measurements, allowing for repeated data collection throughout multiple sample injections. Sample injections are repeated until sufficient neutron scattering statistics are collected. The mixing setup can be programmed to systematically vary conditions to measure the kinetics at different mixing ratios, sample concentrations, additive concentrations, and temperatures. The minimum sample volume required per injection is approximately 150 µL depending on the path length of the quartz cell.
Representative results using this stopped-flow sample environment are presented for rapid lipid exchange kinetics in the presence of an additive, cyclodextrin. The vesicles exchange outer-leaflet (exterior) lipids on the order of seconds and fully exchange both interior and exterior lipids within hours. Measuring lipid exchange kinetics requires in situ mixing to capture the faster (seconds) and slower (minutes) processes and extract the kinetic rate constants. The same sample environment can also be used to probe molecular exchange in other types of liquid samples such as lipid nanoparticles, proteins, surfactants, polymers, emulsions, or inorganic nanoparticles. Measuring the nanoscale structural transformations and kinetics of exchanging or reacting systems will provide new insights into processes that evolve at the nanoscale.
Introduction
Small-angle neutron scattering (SANS) provides a unique way to measure the sizes, shapes, interactions, and organization of various materials on length scales from ≈1 nm to ≈100 nm1,2,3. Recent instruments, including VSANS (very small-angle neutron scattering) instruments with focusing mirrors, push the limits toward measuring even larger length scales up to ≈1000 nm4,5. In general, the unique scattering contrast inherent to neutron scattering methods offers several advantages in measuring the time-evolution of nanoscale structures, such as the aggregation of components in pharmaceutical formulations6, crosslinking and gelation reactions in polymer systems7,8, in meso crystallization of membrane proteins9,10, degradation and unfolding of proteins11,12, and growth of silica-based materials13,14,15. The unique scattering contrast makes time-resolved SANS (TR-SANS) a useful complement to other stopped-flow-based measurements.
Stopped-flow mixing methods often are implemented in small-angle X-ray scattering (SAXS)16,17,18,19,20,21, fluorescence spectroscopy22,23,24,25,26, and light scattering27,28,29,30,31,32 experiments to study kinetic processes on the millisecond time scales. An important difference between SANS and SAXS is that neutron scattering is a nondestructive characterization technique, and as such, SANS can be used to measure the same sample for hours or even days without ionizing radiation damage to the sample, which can happen during higher-flux X-ray scattering experiments33. As repeated SANS measurements will not alter the chemical structure of the probe molecule or sample, the time-evolution can be studied without effects of photobleaching, for example, which can complicate kinetics measurements that rely on fluorescence23,24. Moreover, SANS can be used to measure highly concentrated and optically opaque samples that are often difficult to characterize with light-based techniques such as dynamic light scattering.
In addition to providing structural information on the nanoscale, SANS can be used to probe the local composition of these structures through the variation in neutron scattering length density contrast. The scattering length density (SLD) of different elements varies randomly across the periodic table and varies with different isotopes of the same element. A commonly exploited example is hydrogen (1H or H) and deuterium (2H or D), which have vastly different neutron scattering lengths. Therefore, hydrogen-rich materials, such as surfactants, lipids, proteins, RNA, DNA, and other polymers, can be distinguished from deuterated solvents using SANS without significantly changing the physical properties of the system. However, it is important to note that H/D exchange can affect the density, hydrogen-bonding, and phase transition temperatures in the sample. Nevertheless, the unique sensitivity of SANS to hydrogen-rich materials is especially useful in soft matter research where the samples of interest have lower scattering contrast and signal in X-ray-based techniques such as SAXS. Isotopic substitution also makes SANS a powerful tool for studying molecular exchange kinetics in hydrogen-rich materials by simply mixing H-labeled and D-labeled molecules. Isotopic substitution is particularly useful in systems where bulky fluorescent dyes are larger than the surfactant or lipid molecules of interest and can influence the exchange kinetics34,35.
Time-resolved SANS measurements are advantageous because the measured intensity is a function of time, length scale, and SLD contrast. As such, TR-SANS experiments can be designed to probe the time-dependent changes in the spatial distributions and the compositions of the samples. These unique advantages of SANS have led to important insights into kinetic processes in many soft material systems such as surfactants36,37,38, emulsions39,40,41, lipids34,42,43,44,45,46,47,48,49,50, and polymers51,52,53,54,55,56,57,58,59,60,61,62. Most TR-SANS studies have focused on time scales of minutes to hours. However, many kinetic processes of interest occur on the second time scale and are essential for understanding the underlying mechanisms. Capturing these early time points requires that the solutions be rapidly mixed and measured in situ, in which the mixing is synced with data collection during stopped-flow light scattering27,28,29,30,31,32, fluorescence22,23,24,25,26, and X-ray16,17,18,19,20,21 experiments. This work describes the use of a sample environment designed to rapidly mix multiple liquid samples and inject the mixture into a quartz glass cell for TR-SANS measurements. The mixing device is an adaptation of the recently developed capillary rheoSANS device63 and uses multiple syringe pumps and valves to control the sample mixing and to automate cell cleaning. By connecting syringe pumps to a series of flow selector valves, multiple inlet streams can be repeatedly mixed, measured, rinsed, and dried to facilitate TR-SANS measurements on the seconds time scale.
The current procedure assumes that the samples of interest have been identified and prepared. We focus on the in situ mixing setup and methods to collect TR-SANS data. Neutron scattering data were collected on the VSANS instrument at the NIST Center for Neutron Research (NCNR); however, the procedure should be applicable to other SANS instruments. Readers interested in implementing similar protocols on other SANS instruments should consult with the local instrument scientists to determine the optimal instrument configuration to maximize neutron flux at the desired length scale and time scale most relevant to the kinetic processes of interest. The data presented here were collected using the high flux 'white beam' configuration on VSANS to maximize neutron counts at the loss of spatial resolution5. The detector carriages were positioned to cover a range of scattering vectors (q), 0.005 Å-1 < q < 0.5 Å-1, corresponding to length scales of ≈130 nm to ≈13 nm. The scattering vector is defined as q = 4π/λ sin (θ/2) in which λ is the neutron wavelength, and θ is the scattering angle.
The stopped-flow mixing device used for the TR-SANS measurements consists of multiple pumps, rinsing syringes, sample syringes, flow selectors, as well as adynamic mixer, sample cell, and mixed sample container, as shown in Figure 1. All sealed fluid paths are located inside an air-conditioned enclosure, which includes the syringes, valves, connection tubing, dynamic mixer, and sample cells. A programmable thermoelectric air conditioner is used to control the enclosure temperature in the range from 10 °C to 50 °C within ±1 °C. Note that some of the enclosure insulation was removed to show the working parts of the device. The main mixing device enclosure is positioned on a translational stage on the NG3 VSANS beam line at the NCNR. The enclosure position is adjusted using the translation stage to position the sample cell in the path of the neutron beam (yellow dashed line).
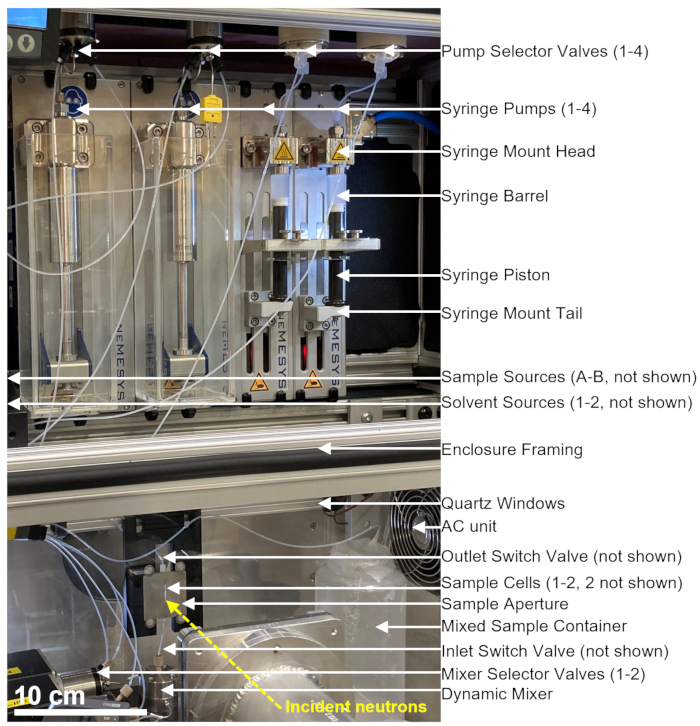
Figure 1: An example setup for combining stopped-flow mixing and small-angle neutron scattering measurements at the VSANS beamline at the NIST Center for Neutron Research. The setup contains four syringe pumps, two syringes for solvent rinsing and two syringes for sample injection, four pump selector valves, two mixer selector valves, a dynamic mixer, a flow-through quartz cell, and a mixed sample container. Incident neutrons scatter off the mixed sample located inside the sample cell. An insulated enclosure with quartz windows and a thermoelectric air-conditioned unit is used to control the sample and all equipment at a constant temperature. The yellow dashed line shows the neutron beam path. Scale bar = 10 cm. Please click here to view a larger version of this figure.
The device depicted in Figure 1 is configured with two sample syringes, two rinsing syringes, and one sample cell. Corresponding flow diagrams for the different steps of the protocol are illustrated in Figure 2. The desired volumes of the two different samples are injected into the mixer and the sample cell (Figure 2A). Once the sample cell is filled, the Inlet Switch Valve (ISV) and Outlet Switch Valve (OSV) are closed to isolate the sample cell from the dynamic mixer and to prevent sample back diffusion into the cell during TR-SANS data collection (Figure 2B). Before the dynamic mixer, the connection tubing varies in length from 10 cm to 1 m and does not affect the mixing delay time. However, tubing connections between the dynamic mixer and the sample cell will affect the mixing delay time and the required sample injection volume. Precut stainless steel tubing with 0.04 inch (1 mm) inner diameter and 100 mm length are used to connect the dynamic mixer, the Mixer Selector Valves (MSV1 and MSV2), and the ISV and OSV. Fluorinated tubing with 1 mm inner diameter and 115 mm length is used to connect the ISV and OSV (or the dynamic mixer outlet) to the sample cell. The total void volume that influences the mixing delay time includes the mixer void volume (0.15 mL), the tubing between the mixer outlet and the sample cell inlet (0.09 mL), and the sample cell volume (0.16 mL). In this example, the total void volume is 0.4 mL. The internal void volumes of valves are negligible compared to the tubing, mixer, and sample cell void volumes. For example, the employed low-pressure selector valves (0.75 mm bore diameter) contain approximate void volumes of 4 µL, while the high-pressure selector valves and switch valves (0.25 mm bore diameter) contain approximate void volumes of 0.5 µL.
After the TR-SANS measurement is complete, the sample is pushed out of the cell with solvent, and rinse solvent is repeatedly pumped through the cell to remove the residual sample and clean the sample cell (Figure 2C). Note that the rinse syringes are connected to larger solvent reservoirs (e.g., water and ethanol) via pump selector values to ensure that adequate solvent volumes are available to clean the sample cell between measurement runs. Solvent sources, sample sources, and mixed sample containers that contain flammable liquids are positioned in a separate enclosure with no electrical equipment to eliminate all possible ignition sources. In addition, vapor-locking bottle caps are used to minimize flammable vapors and solvent evaporation. Finally, the sample cell is dried with a nitrogen gas stream to remove the residual rinse solvent (Figure 2D). The inlet nitrogen gas pressure to the mixer selector valve is regulated to approximately 2 bar (0.2 MPa, gauge pressure) using a manual pressure regulator located on the nitrogen gas cylinder. Once the sample cell is sufficiently cleaned and dried, a newly mixed sample is injected into the sample cell for the next measurement cycle (repeating the mixing and injection illustrated in the flow diagram in Figure 2A).
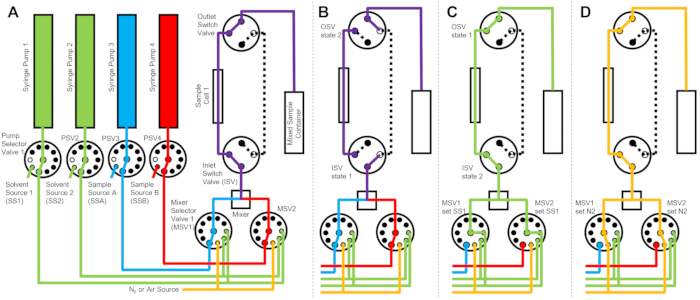
Figure 2: Example flow diagram using one sample cell, two samples mixing, and two rinse solvents for cleaning. (A) Mixing of sample A (blue) and sample B (red), and then flowing the mixed sample (purple) into the sample cell. (B) During data collection, the stopped-flow device state where the ISV and OSV switch valves are closed to isolate the sample cell and prevent back diffusion of the sample during data collection. (C) The cleaning steps where the sample cell is rinsed with rinse solvent from SS1 (green) after data collection. (D) Drying step where the sample cell is dried with nitrogen gas (orange). Abbreviations: PSV = pump selector valve; MSV = mixer selector valve; OSV = outlet switch valve; ISV = inlet switch valve; SS1 = solvent source 1; SSA = sample source A; N2 = nitrogen gas source. Please click here to view a larger version of this figure.
Figure 3 shows flow diagrams for a slightly different version in which the mixing setup is configured with two separate sample cells connected to the same switch valves (Figure 3A). While TR-SANS data are collected in Sample Cell 1, Sample Cell 2 is rinsed (Figure 3B) and dried (Figure 3C). When the data collection is complete for Sample Cell 1, the Inlet Switch Valve directs a newly mixed sample into Sample Cell 2 for data collection (Figure 3D). While TR-SANS data are collected in Sample Cell 2, Sample Cell 1 is rinsed and dried (Figure 3E). This alternating, parallel process between two sample cells minimizes the time between subsequent sample injections and maximizes the use of neutron beam time.
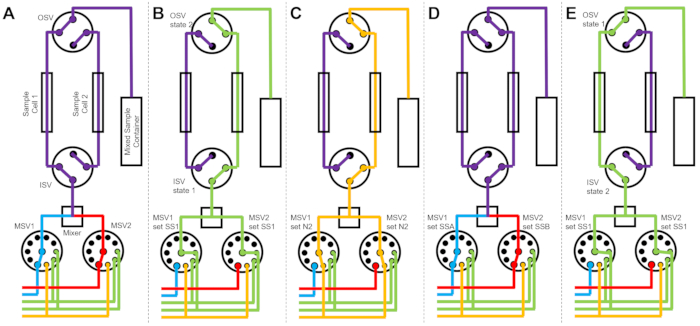
Figure 3: Example flow diagram using a two-sample cells, two samples mixing, and two rinse solvents for cleaning. (A) Mixing sample A (blue) and sample B (red) and then flowing the mixed sample (purple) into sample cell 1. (B) The stopped-flow device state during data collection on sample cell 1 while sample cell 2 is rinsed with solvent from SS1 (green). (C) The stopped-flow device state during data collection on sample cell 1 while sample cell 2 is dried with nitrogen gas (orange). (D) Once data collection of sample cell 1 is complete, a new sample (purple) is immediately mixed and flowed into sample cell 2. (E) The stopped-flow device state during data collection on sample cell 2 while sample cell 1 is rinsed with solvent from SS1 (green). While one sample cell is being measured, the other sample cell is being cleaned and dried. The stopped-flow measurement process alternates between two sample cells to minimize the time between subsequent sample mixing injections. Abbreviations: PSV = pump selector valve; MSV = mixer selector valve; OSV = outlet switch valve; ISV = inlet switch valve; SS1 = solvent source 1; SSA = sample source A; N2 = nitrogen gas source. Please click here to view a larger version of this figure.
A step-by-step protocol is described below for connecting the pumps and tubing lines, priming the system, rinsing and drying the sample cell, and injecting the mixed sample. Although the single-cell configuration is demonstrated for simplicity (Figure 2), the flexible modular setup, protocol, and scripts can be easily modified to implement more syringe pumps, valves, mixers, or sample cell configurations, such as the two-sample cell configuration shown in Figure 3. Representative raw neutron count rate data collected throughout mixing and cleaning injection cycles are shown in Figure 4, while lipid exchange kinetics measured at 3 different temperatures and the extracted normalized scattered intensity corresponding to the fraction of lipids exchanged are shown in Figure 5 and Figure 6, respectively.
Protocol
1. Set up and initiate the stopped-flow system.
- Turn on all pump power supplies and dynamic mixers using the power switch.
- Initiate all pumps and valves in the stopped-flow system control graphical user interface (GUI) by entering the device configuration path and using the commands bus=qmixbus.Bus(), bus.open(), bus.start(), pump=qmixpump.Pump(), pump.enable(), and valve=ViciMultiposSelector() (see example initiation code available in an online open-source repository64).
- Calibrate the pumps before attaching syringes using the command pump.calibrate().
- Confirm that the valves are initiated and move to the correct selector port on command using the command valve.setPort() and valve.getPort().
- Assign the syringe type to be used for each pump using the command pump.set_syringe_param(A, B), in which A is the syringe barrel inner diameter (mm), and B is the syringe max piston stroke distance (mm).
- Connect the sample syringes to the syringe pumps.
- After ensuring that the pumps have been calibrated, screw in clean syringe barrels to the connection at the top of the pump (syringe mount head).
- When using glass syringes, ensure the syringe mount head is loosened before dispensing the fill volume, so that the glass syringe does not break due to excessive force from the syringe piston.
- Screw in the syringe piston to the connection at the bottom of the pump (syringe mount tail).
- After the syringe barrel and syringe piston are connected to the pump, dispense the fill volume of the syringe type using the command pump.empty(), which moves the syringe piston to the top of the syringe barrel.
- When using glass syringes, tighten the syringe mount head after the piston movement stops.
- Connect the tubing to the sample and solvent sources, syringes, valves, mixers, sample cells, and mixed sample container.
- Connect the syringe pump tubing to the pump selector valves.
- Connect the pump selector valve tubing to the sample sources.
- Connect the pump selector valve tubing to the rinsing solvent sources.
- Connect the pump selector valve tubing to the mixer selector valve tubing.
- Connect the mixer selector valve tubing to the nitrogen gas source.
- Connect the mixer selector valve tubing to the mixer inlets.
- Connect the mixer outlet to the inlet switch valve.
- Connect the inlet switch valve to the sample cell inlet.
- Connect the sample cell outlet to the outlet switch valve.
- Connect the outlet switch valve to the mixed sample container.
- Define all tubing and valve connections made (Step 1.7) in the stopped-flow system control GUI by typing in the corresponding port number connections made to each valve (see example control code in the online open-source repository64).
- Calculate the void volume of the tubing between the mixer inlet and sample cell outlet, which defines the minimum amount of sample needed to fill the sample cell for each measurement.
2. Load the sample.
- Set the desired sample fill volume and solvent fill volume in the stopped-flow system control GUI by typing the desired numbers (see example control code in the online open-source repository64).
- Use the pump.aspirate() command to pull in (aspirate) the desired sample and solvent volumes from their sources into the sample syringes through the pump selector valves.
NOTE: When first loading an empty syringe, air will be present at the top of the syringe that must be purged to prime the system with sample and solvent in step 3.
3. Prime the system.
- Use the pump.dispense() command to push out (dispense) all air from syringes, tubing lines, and valves. Ensure that enough liquid volume is dispensed from each syringe to completely remove all air from the syringes, tubing, and valves. If air bubbles are visible inside any tubing, continue dispensing solvent or sample until the bubbles are removed.
- Once air has been purged from the system, perform at least one sample injection and rinsing procedure (without neutron scattering data collected).
- Click to select the cell labeled Start mixing experiment in the control GUI.
- With this cell actively selected, click on the Run button (right triangle) located at the top of the control GUI, or press the Shift and Enter keys together on the keyboard.
- Visually inspect the sample cell to confirm that air bubbles are not present.
- If air bubbles are present, repeat protocol steps 3.1 and 3.2 to further purge any air from the tubing lines.
- If air bubbles are not present in the sample cell, proceed to step 4 to define the remaining experiment protocol steps.
4. Define the stopped-flow mixing protocol in the program script (see code example in the online open-source repository64).
- Enter the temperature set point of the programmable air conditioner (AC) unit that controls the temperature of the insulated enclosure surrounding the stopped-flow device.
- While holding the star button on the AC control unit, press the up and down arrows to change the setpoint temperature. Alternatively, type the desired temperature setpoint in the control GUI and click on Run.
- Wait for 15-30 min to allow the enclosure interior to equilibrate at the desired temperature before starting the kinetic experiments.
NOTE: The accessible temperature range is currently between 10 °C and 50 °C, and the temperature stability is ± 1 °C.
- Enter all rinsing sequence steps by typing in the appropriate volumes, flow rates, times, and number of repetitions into the control GUI.
- Define the volume of each sample to be injected, which defines the total flow rate (Q).
- Define the volume of each solvent to be injected during the rinsing procedure.
- Define the drying time between each rinsing substep (tdry).
- Define the number of rinsing substeps.
- Define the different solvents for the subsequent rinsing steps.
- Define the number of rinsing repetitions to perform after each measurement (nrinse).
- Define the time to fully dry the sample cell and mixer, providing a clean sample cell for the subsequent sample injection (tdry_final).
- Define all sample injection sequence steps by typing the appropriate volumes, flow rates, and times into the stopped-flow system control GUI.
- Define the volume of each sample to be injected and the flow rate.
- Calculate the delay time (tdelay) from the void volume (Vvoid) and the total flow rate (tdelay = Vvoid/Q).
NOTE: The delay time is the time needed to fill the sample cell with the mixed sample. - Define the desired acquisition time for the TR-SANS data such that the entire kinetic process of interest has occurred (tscatt).
- Set the wait time between the end of the scattering experiment and the beginning of the rinse cycles (twait).
NOTE: This wait time should be at least 100 s if the sample neutron transmission is to be measured before it is rinsed from the cell. The sample transmission is needed during data processing to reduce the data to absolute intensity. - Define the number of injection cycles to perform with rinsing sequences run between each injection that are defined in step 4.2 (ncycle).
- Calculate the total time of a single stopped-flow data collection cycle (tcycle) using equation (1).
tcycle = nrinse × (tdelay + tdry) + tdry_final + tdelay + tscatt (1)
in which nrinse = number of rinsing repetitions (step 4.2.6); tdelay = delay time of the stopped flow device (step 4.3.2); tdry = drying time between each rinsing substeps (step 4.2.3); tdry_final = time to fully dry the sample cell and mixer (step 4.2.7); tscatt = desired TR-SANS data acquisition time (step 4.3.3)
5. Define the small-angle neutron scattering parameters in the SANS instrument control GUI.
- Determine the length scales and q-range of interest for each sample.
- Define the instrument configuration to cover the desired q-range of interest, while maximizing the neutron flux at the sample.
- Set the total VSANS data acquisition time in the SANS instrument control GUI to the calculated cycle time in step 4.4 (neutron scattering data acquisition time = tcycle).
- Set the sample transmission measurement time to 100 s in the SANS instrument control GUI.
- Using the SANS instrument control GUI, turn on event mode data collection by typing GenerateEventModeData true in the command line.
6. Collect standard scattering measurements for data reduction before beginning the stopped-flow experiment to process the TR-SANS data.
- Measure the background scattering.
- Ensure that the local instrument shutter is closed.
- Attach the blocked beam sample to the back of the sample aperture, secure the local instrument environment, and open the local instrument shutter.
- Define the blocked beam scattering data acquisition time in the software, and collect blocked beam scattering data, counting for the same duration as the longest scattering data acquisition time (tscatt).
- Once the data collection is complete, close the instrument shutter, and remove the blocked beam sample from the sample aperture.
- Measure the empty cell scattering.
- Ensure that the sample cell has been thoroughly rinsed and dried.
- Open the local instrument shutter.
- Collect the empty cell transmission measurement for 100 s.
- Collect the empty cell scattering measurement, counting for at least the longest acquisition time (tscatt).
7. Begin the stopped-flow experiment.
- Start VSANS scattering data collection in event mode.
- Ensure that the local instrument area is secure and then open the local instrument beam shutter.
- Begin SANS data collection using the SANS instrument control software on the instrument computer by dragging and dropping the desired runs into the instrument queue.
NOTE: To ensure that the earliest time points are measured, begin data collection before starting the stopped-flow mixing experiment. The data will be post-processed at a later step to account for the delay time (tdelay).
- Start the stopped-flow mixing experiment in the control GUI.
- Select the notebook cell labeled Start mixing experiment in the stopped-flow system control GUI.
- With this cell actively selected, click on the Run button (right triangle) located at the top of the stopped-flow device control program, or press the Shift and Enter keys together on the keyboard.
- Confirm that the stopped-flow mixing protocol defined in section 4 has started operating.
- Add the 100 s transmission measurement to the VSANS instrument queue after the scattering run using the SANS instrument control GUI.
- Add one scattering measurement run and one transmission measurement run to the instrument queue for each remaining stopped-flow mixing cycle (ncycle - 1, step 4.3.5) in the SANS instrument control GUI.
8. Process and reduce data to remove all backgrounds, correct for detector sensitivity, and correct for sample transmission.
- Download the scattering data files and associated event files from the server.
NOTE: Separate detector event files will be generated after each VSANS measurement, one event file for each active detector carriage (e.g., front, middle, and/or back detector). - Bin the scattering data to desired time bins using the command events=Rebin(filename) followed by the command events.do_rebinning(timebins), in which the input filename corresponds to the name of the desired SANS data file, and timebins is a list of the desired time bin boundaries in seconds.
NOTE: If the input timebins is entered as a single number instead of a list, then the data will be binned into N number of bins with equal time widths, where N is the input timebins (see software scripts provided by the beamline and available online open-source repository64). - Reduce the binned scattering data using the software provided the beamline65.
9. Analyze the TR-SANS data.
- Calculate the process time of interest (tprocess) from the measurement times using equation (2).
tprocess = ti - tstop + tdelay (2)
Where ti is the measurement time bins starting after the flow is stopped, tstop is the measurement time immediately when the flow is stopped, and tdelay is the delay time. - Plot the q-dependent intensity I(q) as a function of the process time using the time bins defined in step 8.2 and the tprocess calculated in step 9.1.
NOTE: The earliest accessible process time is restricted by tdelay. To measure earlier process time points, increase the total flow rate (Q) or decrease the total void volume (Vvoid). - Extract the kinetic parameters of interest from the change in I(q) as a function of process time.
10. End the experiment.
- Turn off the neutron beam by closing the local instrument shutter.
- Perform a radiation check using a radiation monitor provided by the beamline before disconnecting any parts, tubing, or unloading any samples or mixed sample containers.
- Transfer the syringes, tubing, and mixed sample container to the health physics department.
- Fill out health physics forms and wait for evaluation by health physics personnel.
Results
The representative neutron data shown here measure lipid exchange kinetics in the presence of methyl-β-cyclodextrin (mβCD), an additive that catalyzes the lipid exchange between vesicles with the exchange rate (ke)66,67. Previous fluorescence studies have shown that ke depends on the mβCD concentration, and the half-life of the exchange process is on the order of minutes68. The present experiments use s...
Discussion
The current procedure describes the mixing device and the steps to perform stopped-flow TR-SANS measurements. The device and protocol are optimized for low-viscosity liquid samples where the time scales of interest are ≈1 s to 5 min. For time scales greater than 5 min, manually mixing the samples and loading them into standard scattering cells may be easier and desirable, especially for high-viscosity samples, gels, or pastes. Accessing time scales less than 1 s requires a different mixing apparatus, lower total vo...
Disclosures
The authors have no conflicts of interest to declare.
Acknowledgements
Access to the NG3 VSANS was provided by the Center for High-Resolution Neutron Scattering, a partnership between the National Institute of Standards and Technology and the National Science Foundation under Agreement NO. DMR-2010792. M.H.L.N acknowledges the funding provided by Mitacs Globalink (Canada). The identification of any commercial products or trade names is to foster understanding and does not imply endorsement or recommendation by the National Institute of Standards and Technology.
Materials
| Name | Company | Catalog Number | Comments |
| Dynamic mixer | Analytical Scientific Instruments | 462-0150A | Magnetically coupled rotor, binary dynamic mixer assembly (ternary type available), 0.15 mL dead volume (larger dead volume available) |
| Fluoropolymer tubing | IDEX Health & Science | 1507L | PFA Tubing Natural 1/16 inch OD x 0.040 inch ID x 50 ft |
| Fluoropolymer 1/4-28 flangeless fittings | IDEX Health & Science | XP-245 | PFA flangeless fitting with ferrules, 1/4-28 threading, 1/16 inch OD tubing |
| Glass syringes | Hamilton Company | 81660 | Hamilton 1000 series syringes, 10 mL (81660), model 1010 C syr, 1/4"-28 thread termination, other volumes available |
| High-pressure flow selector valves | Vici Valco | C85X-1570EUTB | Vici 10 position selector valves, 15000 psi max, 0.25 mm bore, 1/16 inch OD tubing, 10-32 coned threaded ports, USB universal actuator |
| High-pressure switch valves | Vici Valco | C82X-1574EUHB | Vici 4 port switch valves, 15000 psi max, 0.25 mm bore, 1/16 inch OD tubing, 10-32 coned threaded ports, USB universal actuator |
| High-pressure syringes | Cetoni | A2019000358 | 3 mL stainless steel syringe, 510 bar max, 21 mL/min flow rate max |
| Low-pressure flow selector valves | Vici Valco | C25-3180EUHB | Vici 10 position selector valves, max 250 psi liquid, 0.75 mm bore, 1/16 inch OD tubing, 1/4-28 threaded ports, USB universal actuator |
| neMESYS high-pressure syringe pumps | Cetoni | A3921000103 | Max force 2600 N |
| neMESYS mid-pressure syringe pumps | Cetoni | A3921000131 | Max force 1000 N |
| Power supply | Cetoni | A3921000127 | Base 600, supplies power for up to 4 high pressure pumps |
| Quartz flow-through sample cell | Starna Scientific | 3-2.30-Q-1/TC | Quartz micro flow cells, 2 mm path length (1 mm available), 2 mm by 2 mm by 30 mm internal dimension |
| Quartz windows | Technical Glass Products | NA | GE 124 Clear fused quartz ground and polished plates, 11.75 inch by 23.75 inch by 0.375 inch thick |
| Stainless steel 10-32 coned compression fittings | IDEX Health & Science | U-321X, U-320X | 316 stainless steel ferrule (U-321X) and nut (U-320X) -Valco type, 10-32 coned, for 1/16 inch OD stainless steel tubing |
| Stainless steel tubing | IDEX Health & Science | U-102 | Stainless Steel Tubing 1/16 inch OD x 0.020 inch ID, 10 cm, various precut lengths available |
| Syringe pump control software | Cetoni | T6000000004 | QmixElements software for nemesys pumps, QmixSDK software development kit |
| Thermoelectric air conditioner | EIC Solutions | AAC-140C-4XT-HC | Thermoelectric air conditioner mounted on insulated enclosure to control the pump, valve, mixer, and sample temperature |
| T-slot railing | McMaster-Carr | 47065T103 | Aluminum t-slotted railing (1.5 inch by 1.5 inch) cut to various lengths |
| Vapor locking bottle caps | Cole-Parmer | EW-12018-02 | Four 304 SS port inserts, 1/4"-28 threads, GL45 bottle cap size, PTFE body, SS threads, PP collar |
References
- Melnichenko, Y. B., Wignall, G. D. Small-angle neutron scattering in materials science: Recent practical applications. Journal of Applied Physics. 102 (2), 021101 (2007).
- Grillo, I., Borsali, R., Pecora, R. Small-angle neutron scattering and applications in soft condensed matter. Soft Matter Characterization. , (2008).
- Hollamby, M. J. Practical applications of small-angle neutron scattering. Physical Chemistry Chemical Physics. 15 (26), 10566-10579 (2013).
- Pipich, V., Fu, Z. KWS-3: Very small angle diffractor with focusing mirror. Journal of large-scale research. 1, 31 (2015).
- Barker, J. G., Kline, S., et al. . 2019 NCNR Annual Report, Special Publication (NIST SP). , (2019).
- Gilbert, P. H., et al. Preservative induced polysorbate 80 micelle aggregation. Journal of Pharmaceutical Sciences. 10 (6), 2395-2404 (2021).
- Terashima, T., et al. In situ and time-resolved small-angle neutron scattering observation of star polymer formation via arm-linking reaction in ruthenium-catalyzed living radical polymerization. Macromolecules. 43 (19), 8218-8232 (2010).
- Hashimoto, K., Fujii, K., Nishi, K., Shibayama, M. Ion gel network formation in an ionic liquid studied by time-resolved small-angle neutron scattering. The Journal of Physical Chemistry B. 122 (40), 9419-9424 (2018).
- Conn, C. E., et al. Membrane protein structures in lipid bilayers; small-Angle neutron scattering with contrast-matched bicontinuous cubic phases. Frontiers in Chemistry. 8, 619470 (2021).
- van't Hag, L., et al. Protein-eye view of the in meso crystallization mechanism. Langmuir. 35 (25), 8344-8356 (2019).
- Mahieu, E., et al. Observing protein degradation by the PAN-20S proteasome by time-resolved neutron scattering. Biophysical Journal. 119 (2), 375-388 (2020).
- Ibrahim, Z., et al. Time-resolved neutron scattering provides new insight into protein substrate processing by a AAA+ unfoldase. Scientific Reports. 7 (1), 40948 (2017).
- Hollamby, M. J., et al. Growth of mesoporous silica nanoparticles monitored by time-resolved small-angle neutron scattering. Langmuir. 28 (9), 4425-4433 (2012).
- Blin, J. L., Impéror-Clerc, M. Mechanism of self-assembly in the synthesis of silica mesoporous materials: in situ studies by X-ray and neutron scattering. Chemical Society Reviews. 42 (9), 4071-4082 (2013).
- Impéror-Clerc, M., Grillo, I., Khodakov, A. Y., Durand, D., Zholobenko, V. L. New insights into the initial steps of the formation of SBA-15 materials: an in situ small angle neutron scattering investigation. Chemical Communications. 8, 834-836 (2007).
- Narayanan, T., Rüter, A., Olsson, U. SAXS/WAXS investigation of amyloid-β(16-22) peptide nanotubes. Frontiers in Bioengineering and Biotechnology. 9, 654349 (2021).
- Angelov, B., et al. DNA/Fusogenic lipid nanocarrier assembly: millisecond structural dynamics. The Journal of Physical Chemistry Letters. 4 (11), 1959-1964 (2013).
- Amann, M., et al. Kinetic pathways for polyelectrolyte coacervate micelle formation revealed by time-resolved synchrotron SAXS. Macromolecules. 52 (21), 8227 (2019).
- Varga, Z., Wacha, A., Bóta, A. Osmotic shrinkage of sterically stabilized liposomes as revealed by time-resolved small-angle X-ray scattering. Journal of Applied Crystallography. 47 (1), 35-40 (2014).
- Panine, P., Finet, S., Weiss, T. M., Narayanan, T. Probing fast kinetics in complex fluids by combined rapid mixing and small-angle X-ray scattering. Advances in Colloid and Interface Science. 127 (1), 9-18 (2006).
- Grillo, I. Applications of stopped-flow in SAXS and SANS. Current Opinion in Colloid & Interface Science. 14 (6), 402-408 (2009).
- Gomez-Hens, A., Perez-Bendito, D. The stopped-flow technique in analytical chemistry. Analytica Chimica Acta. 242, 147-177 (1991).
- Patel, J. T., Belsham, H. R., Rathbone, A. J., Friel, C. T. Use of stopped-flow fluorescence and labeled nucleotides to analyze the ATP turnover cycle of kinesins. Journal of Visualized Experiments: JoVE. (92), e52142 (2014).
- Biro, F. N., Zhai, J., Doucette, C. W., Hingorani, M. M. Application of stopped-flow kinetics methods to investigate the mechanism of action of a DNA repair protein. Journal of Visualized Experiments: JoVE. (37), e1874 (2010).
- Raney, K. D., Sowers, L. C., Millar, D. P., Benkovic, S. J. A fluorescence-based assay for monitoring helicase activity. Proceedings of the National Academy of Sciences of the United States of America. 91 (14), 6644-6648 (1994).
- Roder, H., Maki, K., Cheng, H. Early events in protein folding explored by rapid mixing methods. Chemical reviews. 106 (5), 1836-1861 (2006).
- Milon, A., et al. Osmotic swelling of unilamellar vesicles by the stopped-flow light scattering method. Influence of vesicle size, solute, temperature, cholesterol and three α,ω-dihydroxycarotenoids. Biochimica et Biophysica Acta (BBA) - Biomembranes. 859 (1), 1-9 (1986).
- Gast, K., Nöppert, A., Müller-Frohne, M., Zirwer, D., Damaschun, G. Stopped-flow dynamic light scattering as a method to monitor compaction during protein folding. European Biophysics Journal. 25 (3), 211-219 (1997).
- Antoun, A., Pavlov, M. Y., Tenson, T., Ehrenberg, M. M. Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biological Procedures Online. 6, 35-54 (2004).
- Zhu, Z., Armes, S. P., Liu, S. pH-Induced micellization kinetics of ABC triblock copolymers measured by stopped-flow light scattering. Macromolecules. 38 (23), 9803-9812 (2005).
- Ye, J., et al. Comparative study of temperature-induced association of cyclic and linear poly(N-isopropylacrylamide) chains in dilute solutions by laser light scattering and stopped-flow temperature jump. Macromolecules. 41 (12), 4416-4422 (2008).
- Liu, X., et al. Early stage kinetics of polyelectrolyte complex coacervation monitored through stopped-flow light scattering. Soft Matter. 12 (44), 9030-9038 (2016).
- Garman, E. F., Weik, M. X-ray radiation damage to biological samples: recent progress. Journal of Synchrotron Radiation. 26 (4), 907-911 (2019).
- Garg, S., Porcar, L., Woodka, A. C., Butler, P. D., Perez-Salas, U. Noninvasive neutron scattering measurements reveal slower cholesterol transport in model lipid membranes. Biophysical Journal. 101 (2), 370-377 (2011).
- Marquardt, D., et al. 1H NMR shows slow phospholipid flip-flop in gel and fluid bilayers. Langmuir. 33 (15), 3731-3741 (2017).
- Egelhaaf, S. U., Olsson, U., Schurtenberger, P. Time-resolved SANS for surfactant phase transitions. Physica B: Condensed Matter. 276-278, 326-329 (2000).
- Tabor, R. F., Eastoe, J., Grillo, I. Time-resolved small-angle neutron scattering as a lamellar phase evolves into a microemulsion. Soft Matter. 5 (10), 2125-2129 (2009).
- Gradzielski, M., Bergmeier, M., Hoffmann, H., Müller, M., Grillo, I. Vesicle gel formed by a self-organization process. The Journal of Physical Chemistry B. 104 (49), 11594-11597 (2000).
- Lee, Y. -. T., Li, D. S., Pozzo, L. D. Kinetic analysis of ultrasound-induced oil exchange in oil-in-water emulsions through contrast variation time-resolved small-sngle neutron scattering. Langmuir. 35 (47), 15204-15213 (2019).
- Lee, Y. -. T., Pozzo, L. D. Contrast-variation time-resolved small-angle neutron scattering analysis of oil-exchange kinetics between oil-in-water emulsions stabilized by anionic surfactants. Langmuir. 35 (47), 15192-15203 (2019).
- Roger, K., Olsson, U., Schweins, R., Cabane, B. Emulsion ripening through molecular exchange at droplet contacts. Angewandte Chemie International Edition. 54 (5), 1452-1455 (2015).
- Nakano, M., Fukuda, M., Kudo, T., Endo, H., Handa, T. Determination of Interbilayer and Transbilayer Lipid Transfers by Time-Resolved Small-Angle Neutron Scattering. Physical Review Letters. 98 (23), 238101 (2007).
- Nakano, M., et al. Flip-flop of phospholipids in vesicles: kinetic analysis with time-resolved small-angle neutron scattering. The Journal of Physical Chemistry B. 113 (19), 6745-6748 (2009).
- Nguyen, M. H. L., et al. Methanol accelerates DMPC flip-flop and transfer: A SANS study on lipid dynamics. Biophysical Journal. 116 (5), 755-759 (2019).
- Nguyen, M. H. L., et al. Peptide-induced Lipid flip-flop in asymmetric liposomes measured by small angle neutron scattering. Langmuir. 35 (36), 11735-11744 (2019).
- Nguyen, M. H. L., et al. Time-resolved SANS reveals pore-forming peptides cause rapid lipid reorganization. New Journal of Chemistry. 45 (1), 447-456 (2021).
- Xia, Y., et al. Effects of nanoparticle morphology and acyl chain length on spontaneous lipid transfer rates. Langmuir. 31 (47), 12920-12928 (2015).
- Xia, Y., et al. Morphology-induced defects enhance lipid transfer rates. Langmuir. 32 (38), 9757-9764 (2016).
- Maric, S., et al. Time-resolved small-angle neutron scattering as a probe for the dynamics of lipid exchange between human lipoproteins and naturally derived membranes. Scientific Reports. 9 (1), 7591 (2019).
- Nielsen, J. E., Bjørnestad, V. A., Pipich, V., Jenssen, H., Lund, R. Beyond structural models for the mode of action: How natural antimicrobial peptides affect lipid transport. Journal of Colloid and Interface Science. 582, 793-802 (2021).
- Willner, L., Poppe, A., Allgaier, J., Mokenbusch, M., Richter, D. TIme-resolved SANS for the determintioan of unimer exchange kinetics in block copolymer micelles. Europhysics Letters. 55 (5), 667 (2001).
- Lund, R., Willner, L., Stellbrink, J., Lindner, P., Richter, D. Logarithmic chain-exchange kinetics of diblock copolymer micelles. Physical Review Letters. 96 (6), 068302 (2006).
- Lund, R., Willner, L., Richter, D., Dormidontova, E. E. Equilibrium chain exchange kinetics of diblock copolymer micelles: Tuning and logarithmic relaxation. Macromolecules. 39 (13), 4566-4575 (2006).
- Lund, R., Willner, L., Richter, D., Abe, A., Lee, K. S., Leibler, L., Kobayashi, S. Kinetics of block copolymer micelles studied by small-angle scattering methods. in Controlled Polymerization and Polymeric Structures. Advances in Polymer Science. , 51 (2013).
- Choi, S. -. H., Lodge, T. P., Bates, F. S. Mechanism of molecular exchange in diblock copolymer micelles: hypersensitivity to core chain length. Physical Review Letters. 104 (4), 047802 (2010).
- Choi, S. -. H., Bates, F. S., Lodge, T. P. Molecular exchange in ordered diblock copolymer micelles. Macromolecules. 44 (9), 3594-3604 (2011).
- Lu, J., Bates, F. S., Lodge, T. P. Chain exchange in binary copolymer micelles at equilibrium: confirmation of the independent chain hypothesis. ACS Macro Letters. 2 (5), 451-455 (2013).
- Lu, J., Bates, F. S., Lodge, T. P. Remarkable effect of molecular architecture on chain exchange in triblock copolymer micelles. Macromolecules. 48 (8), 2667-2676 (2015).
- Kelley, E. G., et al. Size evolution of highly amphiphilic macromolecular solution assemblies via a distinct bimodal pathway. Nature Communications. 5 (1), 3599 (2014).
- Murphy, R. P., Kelley, E. G., Rogers, S. A., Sullivan, M. O., Epps, T. H. Unlocking chain exchange in highly amphiphilic block polymer micellar systems: influence of agitation. ACS Macro Letters. 3 (11), 1106-1111 (2014).
- Schantz, A. B., et al. PEE-PEO block copolymer exchange rate between mixed micelles is detergent and temperature activated. Macromolecules. 50 (6), 2484-2494 (2017).
- Lantz, K. A., et al. Cavitation enables switchable and rapid block polymer exchange under high-χN conditions. Macromolecules. 51 (17), 6967-6975 (2018).
- Murphy, R. P., et al. Capillary RheoSANS: measuring the rheology and nanostructure of complex fluids at high shear rates. Soft Matter. 16 (27), 6285-6293 (2020).
- Stopped Flow Sans. usnistgov Available from: https://github.com/usnistgov/stopped-flow-sans (2021)
- Kline, S. Reduction and analysis of SANS and USANS data using IGOR Pro. Journal of Applied Crystallography. 39 (6), 895-900 (2006).
- Doktorova, M., et al. Preparation of asymmetric phospholipid vesicles for use as cell membrane models. Nature Protocols. 13 (9), 2086-2101 (2018).
- Huang, Z., London, E. Effect of cyclodextrin and membrane lipid structure upon cyclodextrin-lipid interaction. Langmuir. 29 (47), 14631-14638 (2013).
- Sugiura, T., Ikeda, K., Nakano, M. Kinetic analysis of the methyl-β-cyclodextrin-mediated intervesicular transfer of pyrene-labeled phospholipids. Langmuir. 32 (51), 13697-13705 (2016).
- Scott, H. L., et al. On the mechanism of bilayer separation by extrusion, or why your LUVs are not really unilamellar. Biophysical Journal. 117 (8), 1381-1386 (2019).
- Dicko, C., et al. NUrF-Optimization of in situ UV-vis and fluorescence and autonomous characterization techniques with small-angle neutron scattering instrumentation. Review of Scientific Instruments. 91 (7), 075111 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved