Method Article
Modeling an Enzyme Active Site using Molecular Visualization Freeware
In This Article
Summary
A key skill in biomolecular modeling is displaying and annotating active sites in proteins. This technique is demonstrated using four popular free programs for macromolecular visualization: iCn3D, Jmol, PyMOL, and UCSF ChimeraX.
Abstract
Biomolecular visualization skills are paramount to understanding key concepts in the biological sciences, such as structure-function relationships and molecular interactions. Various programs allow a learner to manipulate 3D structures, and biomolecular modeling promotes active learning, builds computational skills, and bridges the gap between two dimensional textbook images and the three dimensions of life. A critical skill in this area is to model a protein active site, displaying parts of the macromolecule that can interact with a small molecule, or ligand, in a way that shows binding interactions. In this protocol, we describe this process using four freely available macromolecular modeling programs: iCn3D, Jmol/JSmol, PyMOL, and UCSF ChimeraX. This guide is intended for students seeking to learn the basics of a specific program, as well as instructors incorporating biomolecular modeling into their curriculum. The protocol enables the user to model an active site using a specific visualization program, or to sample several of the free programs available. The model chosen for this protocol is human glucokinase, an isoform of the enzyme hexokinase, which catalyzes the first step of glycolysis. The enzyme is bound to one of its substrates, as well as a non-reactive substrate analog, which allows the user to analyze interactions in the catalytic complex.
Introduction
Understanding representations of the molecular world is critical to becoming an expert in the biomolecular sciences1, because the interpretation of such images is key to understanding biological function2. A learner's introduction to macromolecules usually comes in the form of two-dimensional textbook images of cell membranes, organelles, macromolecules, etc. but the biological reality is that these are three-dimensional structures, and an understanding of their properties requires ways to visualize and extract meaning from 3D models.
Accordingly, the development of biomolecular visual literacy in upper division molecular life science courses has gained attention, with a number of articles reporting on the importance and difficulties of teaching and assessing visualization skills1,3,4,5,6,7,8,9. The response to these articles has been an increase in the number of classroom interventions, typically within a semester in a single institution, wherein molecular visualization programs and models are used to target difficult concepts2,10,11,12,13,14,15. Additionally, researchers have sought to characterize how students use biomolecular visualization programs and/or models to approach a specific topic16,17,18,19. Our own group, BioMolViz, has described a Framework that subdivides overarching themes in visual literacy into learning goals and objectives to guide such interventions20,21, and we lead workshops that train faculty to use the Framework in the backward design of assessments to measure visual literacy skills22.
At the center of all of this work is a critical skill: the ability to manipulate structures of macromolecules using programs for biomolecular visualization. These tools were developed independently using a variety of platforms; therefore, they can be rather unique in their operation and use. This necessitates program-specific instructions, and the identification of a program that a user is comfortable with is important to facilitate continued implementation.
Beyond the very basics of manipulating structures in 3D (rotating, selecting, and altering the model), a major goal is to model the active site of a protein. This process allows a learner to develop their understanding in three overarching themes described by the BioMolViz Framework: molecular interactions, ligands / modifications, and structure-function relationships20,21.
Four popular choices of programs for biomolecular visualization include: Jmol/JSmol23, iCn3D24, PyMOL25, and UCSF Chimera26,27. We encourage those new to Chimera to use UCSF ChimeraX, the next generation of the Chimera molecular visualization program, which is the currently supported version of the program.
In this protocol, we demonstrate how to use each of these four programs to model the active site of human glucokinase with a bound substrate analog complex (PDB ID: 3FGU), and to display measurements to illustrate specific binding interactions28. The model represents a catalytic complex of the enzyme. To capture the active site in the pre-catalysis state, a non-hydrolyzable analog of ATP was bound to the glucokinase active site. This phosphoaminophosphonic acid-adenylate ester (ANP) contains a phosphorous-nitrogen bond instead of the usual phosphorous-oxygen linkage at this position. The active site also contains glucose (denoted BCG in the model) and magnesium (denoted MG). Additionally, there is a potassium ion (K) in the structure, resulting from potassium chloride used in the crystallization solvent. This ion is not critical for biological function and is located outside of the active site.
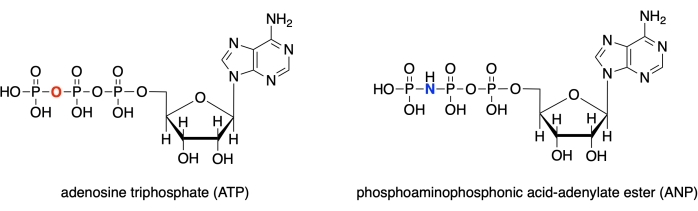
Figure 1: ATP/ANP structures. Adenosine triphosphate (ATP) structure compared to the phosphoaminophosphonic acid-adenylate ester (ANP). Please click here to view a larger version of this figure.
The protocol demonstrates the selection of the bound ligands of the substrate analog complex and the identification of active-site residues within 5 Å of the bound complex, which captures amino acids and water molecules capable of making relevant molecular interactions, including hydrophobic and van der Waals interactions.
The display is initially manipulated to show the majority of the protein in a cartoon representation, with the active site amino acid residues in stick representation to show the relevant atoms of the protein and highlight the molecular interactions. After step 3 of the protocol for each program, these representations have been applied and the view of the protein is similar across programs (Figure 2). At the end of the protocol, the protein cartoon is hidden to simplify the view, and focus on the active site.
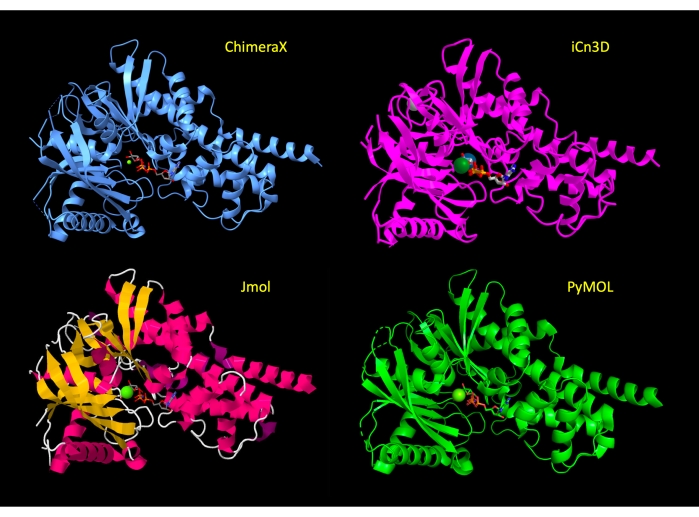
Figure 2: Structure comparison across programs. Comparison of the structure of 3FGU in each program following the Adjust the Representation step (step 2 or 3 of each protocol). Please click here to view a larger version of this figure.
CPK coloring is applied to the active site amino acids and bound ligands29,30. This coloring scheme distinguishes atoms of different chemical elements in molecular models shown in line, stick, ball and stick, and space-filling representations. Hydrogen is white, nitrogen is blue, oxygen is red, sulfur is yellow, and phosphorus is orange in the CPK coloring scheme. Traditionally, black is used for carbon, although in modern use, carbon coloring may vary.
Hydrogen atoms are not visible in crystal structures, although each of these programs is capable of predicting their location. Adding the hydrogen atoms to a large macromolecular structure can obscure the view, thus they are not displayed in this protocol. Accordingly, hydrogen bonds will be shown by measuring from the center of two heteroatoms (e.g., oxygen to oxygen, oxygen to nitrogen) in these structures.
Program Overviews
Downloadable Graphical User Interfaces (GUIs): PyMOL (Version 2.4.1), ChimeraX (Version 1.2.5), and Jmol (Version 1.8.0_301) are GUI-based molecular modeling tools. These three interfaces feature command lines to input typed code; many of the same capabilities are available through menus and buttons in the GUI. A common feature in the command line of these programs is that the user may load and re-execute previous commands using the up and down arrow keys on the keyboard.
Web-based GUIs: iCn3D (I-see-in-3D) is a WebGL-based viewer for interactive viewing of three-dimensional macromolecular structures and chemicals on the Web, without the need to install a separate application. It does not use a command line, though the full web version features an editable command log. JSmol is a JavaScript or HTML5 version of Jmol for use on a website or in a web browser window, and is very similar in operation to Jmol. JSmol can be used to create online tutorials, including animations.
Proteopedia31,32, FirstGlance in Jmol33, and the JSmol web interface (JUDE) at the Milwaukee School of Engineering Center for BioMolecular Modeling are examples of such Jmol-based online design environments34. The Proteopedia wiki is a teaching tool that allows the user to model a macromolecule structure and create pages featuring these models within the website35. The Proteopedia scene authoring tool, built using JSmol, integrates a GUI with additional features not available in the Jmol GUI.
Jmol and iCn3D are based on the Java programming language; JSmol uses either Java or HTML5, and PyMOL and ChimeraX are based on the Python programming language. Each of these programs loads protein data bank files, which can be downloaded from the RCSB Protein Data Bank under a 4-digit alphanumeric PDB ID36,37. The most common file types are Protein Data Bank (PDB) files containing the .pdb extension and Crystallographic Information File (CIF or mmCIF) containing the .cif extension. CIF has superseded PDB as the default file type for the Protein Data Bank, but both file formats function in these programs. There can be slight differences in the way the sequence/structure is displayed when using CIF as opposed to PDB files; however, the files function similarly and the differences will not be addressed in detail here. The Molecular Modeling Database (MMDB), a product of the National Center for Biotechnology Information (NCBI), is a subset of PDB structures to which categorical information has been associated (e.g., biological features, conserved protein domains)38. iCn3D, a product of the NCBI, is capable of loading PDB files containing the MMDB data.
To view a model, the user can download the desired file from the dedicated Protein Data Bank page for the structure (e.g., https://www.rcsb.org/structure/3FGU), and then use the dropdown File menu of the program to open the structure. All of the programs are also capable of loading a structure file directly through the interface, and that method is detailed within the protocols.
The ChimeraX, Jmol, and PyMOL GUIs each contain one or more windows of the console that may be resized by dragging the corner. iCn3D and JSmol are entirely contained in a web browser. When using iCn3D, the user may need to scroll within the pop-up windows to reveal all menu items, depending on screen size and resolution.
The protocols detailed here provide a simple method to display the active site of the enzyme using each program. It should be noted that there are multiple ways to execute the steps in each program. For example, in ChimeraX, the same task may be executed using dropdown menus, the toolbar at the top, or the command line. Users interested in learning a specific program in detail are encouraged to explore the online tutorials, manuals, and Wikis available for these programs39,40,41,42,43,44,45,46.
Existing manuals and tutorials for these programs present the items in this protocol as discrete tasks. To display an active site, the user must synthesize the required operations from the various manuals and tutorials. This manuscript augments existing tutorials available by presenting a linear protocol for modeling a labeled active site with molecular interactions, providing the user with a logic for active site modeling that can be applied to other models and programs.
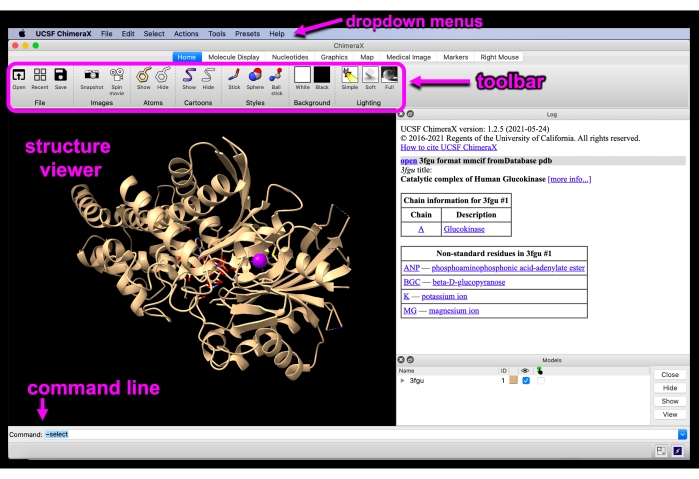
Figure 3: ChimeraX GUI. ChimeraX GUI interface with the dropdown menus, toolbar, structure viewer, and command line labeled. Please click here to view a larger version of this figure.
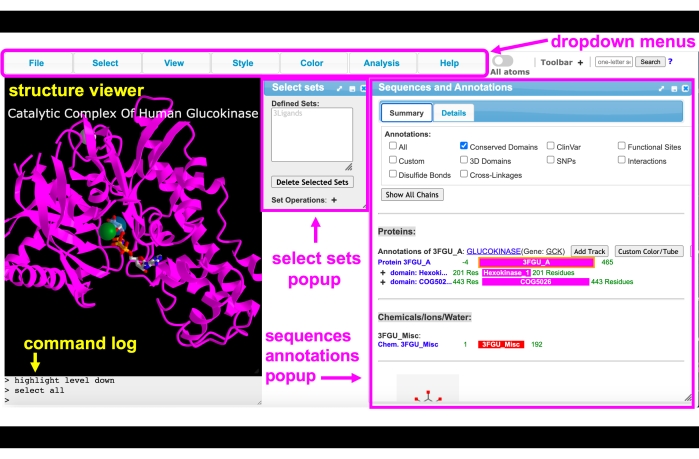
Figure 4: iCn3D GUI. iCn3D GUI interface with the dropdown menus, toolbar, structure viewer, command log, select sets pop-up, and sequence and annotations pop-up menus labeled. Please click here to view a larger version of this figure.
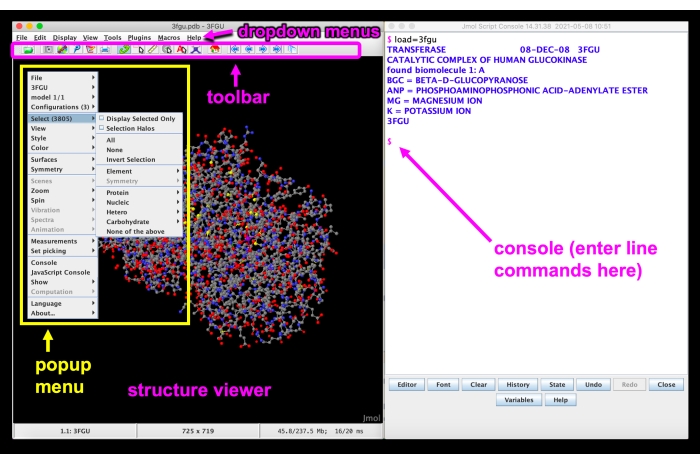
Figure 5: Jmol GUI. Jmol GUI interface with the dropdown menus, toolbar, structure viewer, pop-up menu, and console/command line labeled. Please click here to view a larger version of this figure.
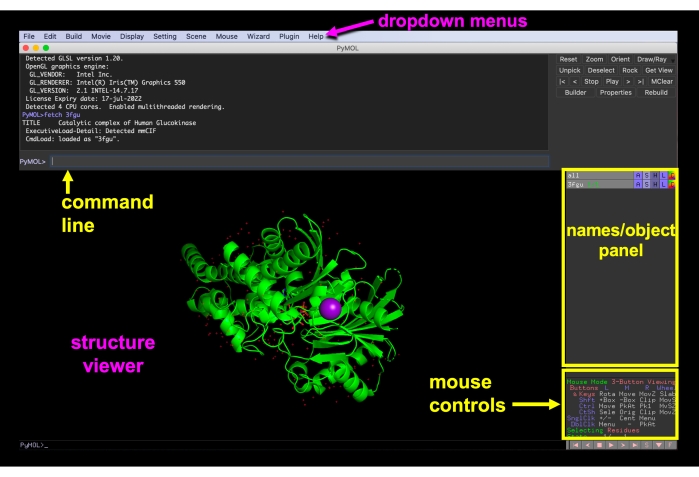
Figure 6: PyMOL GUI. PyMOL GUI interface with the dropdown menus, structure viewer, names/object panel, mouse controls menu, and command line labeled. Please click here to view a larger version of this figure.
Protocol
NOTE: The protocol for each program is outlined in ten overarching steps, (1) Loading the structure into the program, (2) Identifying the ligands in the active site, (3) Adjusting the representation, (4) Selecting residues within 5 Å to define an active site, (5) Showing the interactions of the enzyme with the active site ligands, (6) Displaying the side chains as sticks and showing/adjusting the active site water molecules, (7) Simplifying the structure, (8) Labeling ligands and hydrogen-bonded side chains, (9) Saving the rendering at any point to return to work on it or share with others, (10) Saving an image for embedding or printing. Steps 1, 4, and 7-10 are identical for each protocol; however, due to the unique operation of each program, some protocols are more efficiently executed when steps 2/3 and 5/6 are interchanged.
1. UCSF ChimeraX protocol
NOTE: Trackpad and Mouse Controls. To rotate, click and drag or use two-finger drag (mouse: left click and drag). To zoom, pinch and spread (Mac) or control + two-finger movement (PC) (mouse: scroll wheel). To translate (i.e., move the entire structure) press the option + click and drag (Mac) or shift + click and drag (PC) (mouse: right click and drag). To re-center, use the dropdown menus at the top of the interface to click Actions > View.
- Loading the structure into ChimeraX: In the command line located at the bottom of the GUI that is preceded by "Command:", type:
open 3fgu
NOTE: After inputting any typed line command, press return on the keyboard to execute it. - Identifying the ligands in the active site: Ensure there are two representations, a cartoon ribbon and sticks. Using the mouse, rotate/zoom the protein to best visualize the ligands displayed near the center of the protein, which are shown as sticks. Hover over a ligand to show its name.
- Adjusting the representation: Use the commands in the substeps below to recolor the protein and ligands, apply CPK coloring to non-carbon atoms, and then deselect the selection. Selected parts of the molecule become highlighted in green.
- Use the dropdown menus at the top of the interface to change the coloring: Click on Actions > Color > Cornflower Blue. Then, click on Select > Structure > Ligand. To select the color, click on Actions > Color > Gray. To apply CPK coloring, click on Select > All, and then click on Actions > Color > By Heteroatom. Finally, clear the selection by clicking on Select > Clear.
NOTE: The selection may also be cleared by pressing control and clicking in the black background of the structure viewer or in the command line by typing: ~select. By default, for most structures containing 1-4 chains, ChimeraX will automatically show water molecules and amino acid residues within 3.6 Å of ligands and ions. - Use the dropdown menu to hide the currently displayed atoms by clicking on Actions > Atoms/Bonds > Hide.
- Use the dropdown menu to show the ligands and Mg ion in the active site by clicking on Select > Structure > Ligand. Then, click on Actions > Atoms/Bonds > Show. Next, click on Select > Residues > MG, and then on Actions > Atoms/Bonds > Show. To clear the selection, click on Select > Clear.
NOTE: After making the selection with the dropdown menu, step 1.3.3 may be executed by clicking on the Hide and Show buttons in the Atoms toolbar.
- Use the dropdown menus at the top of the interface to change the coloring: Click on Actions > Color > Cornflower Blue. Then, click on Select > Structure > Ligand. To select the color, click on Actions > Color > Gray. To apply CPK coloring, click on Select > All, and then click on Actions > Color > By Heteroatom. Finally, clear the selection by clicking on Select > Clear.
- Selecting residues within 5 Å to define an active site: In the structure viewer, to select the ligands press control + shift and perform the mouse click on any single atom or bond in each of the three ligands, i.e., BCG, ANP, and Mg.
- Next, press the up arrow key on the keyboard until all the atoms of the three ligands are highlighted with a green glow. Define this selection for future use by clicking in the dropdown menu Select > Define Selector. In the pop-up menu, type:
ligands to name the current selection, and then click on OK.
NOTE: Clicking the up arrow too many times in step 1.4.1 will select the entire protein. In that case, click the down arrow button until only the atoms of the three ligands are selected. - Use the dropdown menu to select the residues within 5 Å of the ligands: Click on Select > Zone. In the pop-up window that appears, toggle the Select dropdown menu to Residues, and ensure that the top box is checked (check the less than (<) distance and set to 5.0 Å). Then, click on OK. Only residues which are less than 5 Å away will be highlighted.
NOTE: Steps 1.4-1.4.2 can be simplified extensively using the command line, by typing:
name frozen ligands :BGC:MG:ANP
select zone ligands 5 extend true residues true
- Next, press the up arrow key on the keyboard until all the atoms of the three ligands are highlighted with a green glow. Define this selection for future use by clicking in the dropdown menu Select > Define Selector. In the pop-up menu, type:
- Displaying the side chains as sticks and showing/adjusting the active site water molecules: Use the dropdown menu to display the selection and center and zoom on the selection by clicking on Actions > Atoms/Bonds > Show to show them. To center the selection, click on Actions > View. Then, to clear the selection, click on Select > Clear or click anywhere in the empty space .
- Showing the interactions of the enzyme with the active site ligands: Use the dropdown menus and click on Select > User-Defined Selectors > Ligands. Then, click on Tools > Structure Analysis > H-bonds. In the pop-up window, ensure that Limit by Selection is checked, the dropdown menu is set to With At Least One End Selected, and Select Atoms is checked, and then click on OK. To clear the selection, click on Select > Clear.
NOTE: Check the Distance Label box to see bond lengths in Å; however, this makes the view very busy. Finally, you may change the color of the H-bonds by clicking on the Color box and selecting a new color in the pop-up window. - Simplifying the structure: using the top cartoon toolbar to hide the cartoon or click on the dropdown menu: Actions > Cartoon > Hide.
- Labeling ligands and hydrogen-bonded side chains: Use the mouse to select residues that are hydrogen bonded to the ligands (connected by the dashed lines), as in step 1.4. Then, in the dropdown menus, click on Actions > Label > Residues > Name Combo. Next, click on Select > User-Defined Selectors > Ligands. Then, click on Actions > Label > Residues > Off. Finally, clear the selection, by clicking on Select > Clear.
- Saving the rendering at any point to return to work on it or share with others: in the dropdown menu click on File > Save. Select a location, enter a filename, and click on Save.
NOTE: Ensure the format is set to: ChimeraX session *.cxs. - Saving an image for embedding or printing: First use the mouse to orient the molecule as desired. Change the background color to white by typing in the command line:
set bgColor white
Finally, click on the snapshot icon in the Toolbar. The image will save to the desktop.
NOTE: Background color is also available in the dropdown menu; on a Mac, click on UCSF ChimeraX > Preferences; on a PC, click on Favorites > Settings > Background.
2. iCn3D protocol
NOTE: Trackpad and Mouse Controls: To rotate, click and drag (mouse: left click and drag). To zoom, pinch and spread (mouse: rotate the scroll wheel). To translate (i.e., move the entire structure) click and drag with two fingers (mouse: right click and drag). To re-center, hover over View in the top dropdown menus, and then click on Center Selection.
- Loading the structure into iCn3D: Navigate to iCn3D Web-based 3D Structure Viewer and type 3FGU into the Input MMDB or PDB ID box to load the file.
- Identifying the ligands in the active site: Hover over Analysis in the dropdown menu, and then click on Seq. and Annotations. The sequences, in this case Proteins and Chemical/Ions/Water, are shown in a stacked table. Scroll down to see the active site ligands ANP, BGC, and Mg listed. In the structure viewer, hover over the ligands in the active site (shown as sticks in the center of the protein cartoon) to view their names.
- Adjusting the representation: No initial adjustments are required for this protocol.
- Selecting residues within 5 Å to define an active site: To select the ligands, use the Select dropdown menu and click on Select on 3D. Ensure that Residue is checked .
- To select the ligands, hold down the ALT button on a PC or the Option button on a Mac, and click on the first ligand (e.g., BCG) using the mouse or trackpad. Then, press control and click on ANP and MG ligands to add them to the selection.
NOTE: The ligands will become highlighted in yellow as they are selected. - Save this selection using the dropdown menu: Click on Select > Save Selection and use the keyboard to input a name in the pop-up window (e.g., 3Ligands), and then click on Save. The Select Sets pop-up window will now appear.
NOTE: If the selection is incorrect, click on Select > Clear Selection. - Select the residues within 5 Å of the ligands: In the dropdown menu, click on Select > By Distance. In the popup menu that appears, change the second item (Sphere with a radius), to 5 Å by typing in the block. Click on the boxed word Display, and then close the window by clicking on the cross sign in the upper right-hand corner.
NOTE: In the pop-up menu that appears in step 2.4.3, leave the first set with the input "selected" and the second set as "non-selected." Note that the atoms/structures within 5 Å become highlighted with a yellow glow when Display is clicked. - Save the 5 Å active site using the dropdown menu: Hover over Select and click on Save Selection, input a name in the popup window using the keyboard (e.g., 5Ang), and click on Save.
- Next create a new selection that combines the two sets (5Ang and 3Ligands): In the Select Sets pop-up menu, ctrl-click (PC) or command-click (Mac) 5Ang and 3Ligands. Click on Select > Save Selection, use the keyboard to type a name (e.g., 5AFull), and then click on Save.
- To select the ligands, hold down the ALT button on a PC or the Option button on a Mac, and click on the first ligand (e.g., BCG) using the mouse or trackpad. Then, press control and click on ANP and MG ligands to add them to the selection.
- Showing the interactions of the enzyme with the active site ligands such as hydrogen bonds: Hover over Analysis in the dropdown menu and click on Interactions. A comprehensive pop-up menu of all noncovalent interactions will appear.
- Uncheck everything except the "Hydrogen bonds" and "Salt Bridge/Ionic" checkboxes. Click on 3Ligands to select the first set and 5Ang for the second set. Click on the boxed text that reads 3D Display interactions. Close the window by clicking on the cross sign in the upper right-hand corner.
NOTE: The Contact/Interactions presumably represent induced dipole-induced dipole interaction, which often makes the display busy. If desired, alter the distance for any type of interaction. - To show hydrogen bonds only, click on 5Afull in the select sets pop-up window. Then, hover over Analysis in the dropdown menu, and then click on Chem. Binding > Show.
- Uncheck everything except the "Hydrogen bonds" and "Salt Bridge/Ionic" checkboxes. Click on 3Ligands to select the first set and 5Ang for the second set. Click on the boxed text that reads 3D Display interactions. Close the window by clicking on the cross sign in the upper right-hand corner.
- Displaying the side chains as sticks and showing/adjusting the active site water molecules: Use the select sets pop-up menu and click on 5AFull. Then, in the dropdown menus, click on Style > Side Chains > Stick. To apply CPK coloring click on Color > Atom. Finally, click on Style > Water > Spheres (if you prefer larger water molecules).
- Simplifying the structure: In the select sets pop-up menu, click on 5AFull. Then, in the dropdown menus, click on View > View Selection (to just see the 5AFull binding site). Next, click on Style > Proteins > Stick (to show the protein chain as stick instead of ribbon).
- To color the carbon atoms of the ligands a contrasting color, click on Chemicals in the Select Sets pop-up window. Then, in the dropdown menu, click on View > View selection. Next click on Select > Select on 3D (ensure "atom" is checked). Using the controls described in step 2.4.1, use the mouse and keyboard to select all carbon atoms in BGC and ANP. Then, in the dropdown menu click on Color > Unicolor > Cyan > Cyan.
- To redisplay the entire active site, use the Select Sets pop-up window to click on 5AFull. Then, in the dropdown menu, click on View > View Selection.
- Labeling ligands and hydrogen-bonded side chains: Use the select sets pop-up window to select Interface_all, and then, in the dropdown menu, click on Analysis > Label > Per Residue & Number.
NOTE: You will have to reselect Per Residue & Number each time you wish to add a label, even though the menu item will already be checked from a previous label. - Saving the rendering at any point to return to work on it or share with others: In the dropdown menu, click on File > Share Link. Copy the short URL (for example: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?r83NqCz41bu7cmcs8) and paste it onto a browser.
- Saving an image for embedding or printing: In the dropdown menu, click on Select > Toggle highlight. Then, click on Style > Background > White. Finally, click on File > Save files > iCn3D PNG Image and choose the desired size.
3. Jmol Protocol
NOTE: Trackpad and Mouse Controls: To rotate, click and drag (mouse: left click and drag). To zoom: scroll vertically using two fingers (mouse: shift + left click + drag vertically). To translate (i.e., move the entire structure) control + alt + click and drag (PC), control + option + click and drag (Mac). To re-center: shift + double click in the empty space of the structure viewer window.
- Loading the structure into Jmol: Use the dropdown menu at the top of the GUI to set up the workspace with the structure by clicking on File > Console. Then, click on File > Get PDB. In the pop-up window, type: 3fgu
Then, click on OK.
NOTE: Alternately, use the Jmol console to load the structure, by typing: load = 3fgu
NOTE: After inputting any typed line command, press return on the keyboard to execute it. - Adjusting the representation: Open the pop-up menu by right-clicking (or control + click) anywhere in the Structure Viewer Window.
- To change the protein to cartoon representation, in the pop-up menu, click on Select > Selection Halos. Next, click on Select > Protein > All. Finally, click on Style > Scheme > Cartoon.
NOTE: Selection halos puts a yellow outline (glow) around all the selected atoms. - Use the top dropdown menu to hide the waters by clicking on Display > Select > Water. Next, click on Display > Atom > None, and finally click on Display > Select > None.
- To change the protein to cartoon representation, in the pop-up menu, click on Select > Selection Halos. Next, click on Select > Protein > All. Finally, click on Style > Scheme > Cartoon.
- Identifying the ligands in the active site: Use the mouse to zoom in on the active site, and then use the commands in the substeps to display the ligands as sticks.
NOTE: Ligand names appear in the Jmol console when you load the file. You can also view bound ligand names using the pop-up menu, by clicking on Select > Hetero > by HETATM.- Hover over the ligands with the mouse to view their names. The active site is near the center of the structure; the ligands MG, BGC, and ANP are located in the active site.
- Select the ligands BCG and ANP: Using the Jmol console, type:
select BGC, ANP - To display the ligands BCG and ANP as sticks, use the pop-up menu and click on Style > Scheme > Sticks.
- Selecting residues within 5 Å to define an active site: In the Jmol console, type the following command to select atoms within 5 Å of the three ligands:
select within (5, (bgc,anp,mg))- To select full amino acid residues, type the following in the console and press Enter
select within(group, selected)
NOTE: The Jmol console is the best way to select the residues within 5 Å.
- To select full amino acid residues, type the following in the console and press Enter
- Displaying the side chains as sticks and showing/adjusting the active site water molecules: Right-click to bring up the pop-up menu, and hover over Style > Scheme > Sticks.
NOTE: Step 3.5 shows the active site side chains in stick representation. There will still be some empty halos in the structure which represent the water molecules in the active site.- In the Jmol console, re-execute the following command:
select within (5, (bgc,anp,mg))
NOTE: To re-execute a command, click within the console, and then use the arrow keys on the keyboard until that command appears, and click on enter to re-execute it. - To display the water molecule atoms, remove the ligands and protein from the selection by typing the following two commands:
select remove group protein
select remove group hetero and not water - To display the water molecules, click on the dropdown menu Display. Hover over Atom and click on 20% van der Waals. The green Magnesium ions will still be shown as sticks. Display the magnesium ion in the more common sphere representation by typing the following commands in the Jmol console:
select Mg
spacefill 50% - Recolor the ligands to distinguish them from the protein: In the Jmol console, type the following to execute a command that recolors the ligands in a lighter color scheme:
select (bgc,anp) and carbon; color [211,211,211]
select (bgc,anp) and oxygen; color [255,185,185]
select (bgc,anp) and nitrogen; color [150,210,255]
select (bgc,anp) and phosphorus; color [255,165,75]
select Mg; color palegreen
- In the Jmol console, re-execute the following command:
- Showing the interactions of the enzyme with the active site ligands: Using the Jmol console, execute each line of the following command:
define ligbind (ANP, BGC, MG)
select within (5, (bgc,anp,mg))
select remove group hetero and not water- To show lines to illustrate hydrogen bonds, type this command in the Jmol console:
connect 3.3 (ligbind and (oxygen or nitrogen)) (selected and (oxygen or nitrogen)) strut yellow
Then, modify the thickness of the lines by typing the following command in the console:
select all; strut 0.1; select none
- To show lines to illustrate hydrogen bonds, type this command in the Jmol console:
- Simplifying the structure: To hide the cartoon of the protein and clear the selection, in the Jmol console, type:
select all; cartoon off; select none - Labeling ligands and hydrogen-bonded side chains: In the pop-up window, click on Set Picking > Select Atom. Click on an atom in one of the hydrogen bonded residues. The amino acid and residue numbers appear in the console. Then, use the console to type a label, for example:
label Glu-256 - Saving the rendering at any point to return to work on it or share with others: In the top menu, click on the camera icon. Type a file name and select a location to save.
NOTE: An exported JPEG file (.jpg) contains the information for both an image as it appears in the display window at the time of exporting, as well as the current state of the model. To reload the model, open Jmol and drag the saved JPEG file into the Jmol Display Window. - Saving an image for embedding or printing: In the Jmol console, recolor the background to white, by typing:
background white
As in step 3.9, click on the camera icon and save the file.
4. PyMOL protocol
NOTE: Trackpad and Mouse Controls: To rotate, click and drag (mouse: left click and drag). To zoom, pinch and spread (mouse: right click and drag). To translate (i.e., move the entire structure), control + click and drag (mouse: command + left click and drag). To re-center go to the right-hand object panel and click on A > Orient or Center.
- Loading the structure into PyMOL: In the command line near the top of the GUI (preceded by "PyMOL>"), type:
fetch 3FGU
NOTE: After inputting any typed line command, press return on the keyboard to execute it. - Adjusting the representation: In the names/object panel on the right-hand side of the PyMOL window, to the right of "3FGU" click on H > Waters.
- Identifying the ligands in the active site: First turn on the sequence viewer by clicking on the top dropdown menu: Display > Sequence.
- Scroll the gray bar to the right until you find the ligand names (BCG, ANP, MG, K).
NOTE: There are two representations, a cartoon ribbon and sticks; the ligands are shown as sticks. Ensure the selecting mode in the mouse controls on the bottom right panel is set to Residue and 3-Button Viewing mode by clicking on these names to toggle through the options. - Using the mouse, rotate and zoom to make the ligands visible.
- Scroll the gray bar to the right until you find the ligand names (BCG, ANP, MG, K).
- Selecting residues within 5Å to define an active site: To select the ligands in the active site, click on each one (BCG, ANP, MG) in the structure viewer. A new selection pops up in the names/object panel; to the right of this new object named "sele", click on the A button, and then click on Rename in the pop-up menu.
NOTE: To clear an undesired selection, click on the empty space in the structure viewer to deselect.- Using the keyboard, delete the letters "sele" that appear on the top left-hand side of the structure viewer window, and in place of them, type:
ligands
NOTE: Steps 4.4-4.4.1 can be done using the command line; type:
sele ligands, resn BGC+ANP+MG - Use this selection to define the area around the ligands by first duplicating it, click on ligands > A > Duplicate. Then, click on sel01 > A > Rename
Using the keyboard, delete the letters "se101" and type:
active - Modify this selection to show residues within 5 Å: In the names/object panel, click on active > A > Modify > Expand > by 5 A, Residues. Then, to show these residues as sticks, click on active > S > Licorice > Sticks. Finally, click in the empty space in the Structure Viewer to clear the selection.
NOTE: Step 4.4.3 can be done using the command line, type:
sele active, byres all within 5 of ligands
show sticks, active
- Using the keyboard, delete the letters "sele" that appear on the top left-hand side of the structure viewer window, and in place of them, type:
- Displaying the side chains as sticks and showing/adjusting the active site water molecules: In the names/object panel click on ligands > A > Duplicate. To rename the selection, click on Sel02 > A > Rename Selection. Delete the letters in the renaming menu that appears in the top right of the structure viewer, and type:
active_water- To adjust the new selection to contain active site water molecules, click on active_water > A > Modify > Around > Atoms Within 4 Angstroms. To modify this further and limit to water molecules, click on active_water > A > Modify > Restrict > To Solvent. Finally, click on active_water > A > Preset > Ball and Stick.
NOTE: The GUI allows selection within 4 Å; line commands allow selection of a more appropriate distance of 3.3 Å for hydrogen bonding water molecules. The van der Waals radii of the spheres cannot be set in the GUI, but the "ball and stick" selection is close to 0.5 Å.
NOTE: Steps 4.5-4.5.1 may be executed using the command line, by typing each line of the following code:
select active_water, ((ligands)around 3.3) and (resn HOH)
show spheres, active_water
alter active_water, vdw=0.5
rebuild
- To adjust the new selection to contain active site water molecules, click on active_water > A > Modify > Around > Atoms Within 4 Angstroms. To modify this further and limit to water molecules, click on active_water > A > Modify > Restrict > To Solvent. Finally, click on active_water > A > Preset > Ball and Stick.
- Showing the interactions of the enzyme with the active site ligands. Zoom in on the active site by clicking on active > A > Zoom. To find the polar contacts between the ligands and active site, click on ligands > A > Find > Polar Contacts > To Any Atoms. Show distances as labels by clicking on ligands_polar_contacts > S > Labels.
- Simplifying the structure: Hide the cartoon of the protein, which hides the part of the protein that is not in the active site, by clicking on 3FGU > H > Cartoon in the names/object panel. Next, hide the labels of the hydrogen bond length by clicking on ligands_polar_contacts > H > Labels in the names/object panel.
- To color the ligands to differentiate them from the protein, click on ligands > C > By Element > CHNOS and select the option where "C" is cyan (a light blue).
NOTE: Step 4.7.1 may be executed using the command line. Type:
color cyan, ligands
color atomic, ligands & !elem C
- To color the ligands to differentiate them from the protein, click on ligands > C > By Element > CHNOS and select the option where "C" is cyan (a light blue).
- Labeling ligands and hydrogen-bonded side chains: In the names/object panel, on the buttons to the right of any object name, click on active > L > Residues.
- Saving the rendering at any point to return to work on it or share with others: In the dropdown menu click on File > Save Session As. Then, select a location in the pop-up window, type a filename, and click on Save.
- Saving an image for embedding or printing: First, change the background to white in the dropdown menu by clicking on Display > Background > White. Export the image as a new file, by clicking on File > Export Image As > PNG.
Results
A successfully executed protocol for each of the programs will result in a molecular model zoomed in on the active site, with active site residues and ligands shown as sticks, the protein cartoon hidden, and ligands displayed with a contrasting color scheme. Interacting amino acid residues should be labeled with their identifiers, and hydrogen bonding and ionic interactions shown with lines. The presence of these features can be determined by visual inspection of the model.
To facilitate this inspection and enable the user to determine whether they have performed the steps of the protocol correctly, we've provided animated figures that present an image of the structure following each step. For ChimeraX, iCn3D, Jmol, and PyMOL, this is illustrated in Figures 7-10, respectively.
Figure 7: ChimeraX protocol output. Animated figure illustrating steps 1.1-1.8 of the ChimeraX protocol. Please click here to download this figure.
Figure 8: iCn3D protocol output. Animated figure illustrating steps 2.1-2.8 of the iCn3D protocol. Please click here to download this figure.
Figure 9: Jmol protocol output. Animated figure illustrating steps 3.1-3.8 of the Jmol protocol. Please click here to download this figure.
Figure 10: PyMOL protocol output. Animated figure illustrating steps 4.1-4.8 of the PyMOL protocol. Please click here to download this figure.
The most common error that can influence the outcome of these protocols is erroneous selection, resulting in part of the structure being displayed in an undesired rendering. This is typically a result of mis-clicking, either on the structure itself, or in one of the display menu buttons. An example of a suboptimal result would be a model containing residues outside of the active site displayed as sticks. The user can begin to analyze if this error has occurred by visually inspecting the residues displayed as sticks and ensuring that they are in the proximity of the active site ligands. An advanced method to evaluate whether or not the displayed residues are within 5Å of the active site ligands is to use the measurement tools built into each program to measure the distance between a nearby ligand and the active site residue. The measurement tools are beyond the scope of this manuscript; however, we encourage interested users to explore the many online tutorials detailing this type of analysis.
We present a specific example of a sub-optimal execution of this protocol, resulting from a mis-click on the names/objects panel in the PyMOL. This error displays the entire protein as sticks, rather than showing only the active site using this representation, as illustrated in Figure 11.
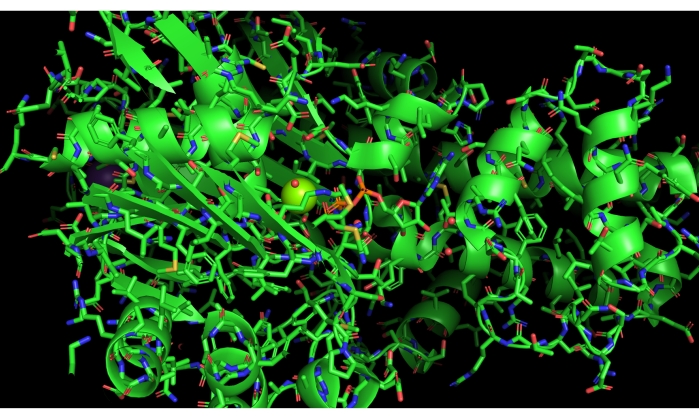
Figure 11: Negative result. Example of a negative result. Mis-selecting the full cartoon in PyMOL and displaying sticks. Please click here to view a larger version of this figure.
To troubleshoot, the user will need to hide the sticks for the entire model (labeled 3FGU in the names/object panel), and then show the stick representation for only the selection named "active," using the hide and show buttons/commands in PyMOL. Recovering the model from this type of error is relatively straightforward once the user is able to create appropriate selections for different parts of the model and display and hide them effectively. It is tempting to restart the protocol and work through the steps another time; however, we encourage the user to not be afraid to go "off script" and experiment with the model. In our experience, working through display errors facilitates progress in understanding the modeling program.
A side-by-side display of the final output from a successfully-executed protocol for each program is shown in Figure 12. The views are oriented similarly to allow the user to compare the appearance of the models created in different programs.
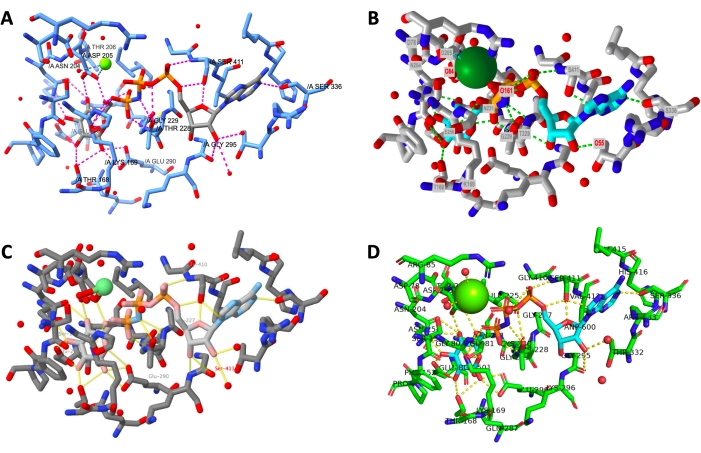
Figure 12: Final structure comparison across programs. Comparison of the structure of each active site rendering at the end of the protocol. A: ChimeraX, B: iCn3D, C: Jmol, D: PyMOL. The PyMOL active site label includes all active site residues and the ligands. The other outputs have only hydrogen bonded side chains labeled. Please click here to view a larger version of this figure.
Discussion
This protocol outlines a ten-step process for the modeling of an enzyme active site, applied to four popular programs for biomolecular modeling. The critical steps of the protocol are: identifying the ligands in the active site, selecting residues within 5 Å to define an active site, and showing the interactions of the enzyme with the active site ligands. Distinguishing the ligands relevant to the biological function is paramount, as this allows the user to define the amino acid residues within 5 Å that can play a role in binding the ligands. Finally, using the program to display molecular interactions allows the user to develop the skills necessary to understand the molecular interactions that promote binding.
A limitation of computer-based molecular modeling protocols is the dependence on specific commands and syntax. While biochemical protocols may be tolerant of small changes in procedure, computer-based investigations may yield wildly different final products if the procedure is not closely adhered to. This is particularly important when using command-line interfaces where program-specific syntax is required to achieve a certain output, and a seemingly insignificant change in punctuation or capitalization can cause a command to fail. There are various Wikis and manuals for each program, where a user can find and troubleshoot command-line inputs; the user should pay careful attention to the details of command syntax. Although most molecular visualization programs include undo commands, due to the complexity of the interfaces, the undo command doesn't always faithfully reverse the last executed step. Therefore, saving the current working state often is encouraged, especially for new users.
Further limitations can arise from the data used to create the model itself. While the standards inherent in the Protein Data Bank ensure a certain level of consistency, users of molecular visualization programs will often encounter unexpected effects in a protein rendering. First, most structures are determined using X-ray crystallography, which provides a single model of the protein; however, NMR structures are often composed of multiple models that can be visualized one at a time. Second, structures determined from crystallography or cryogenic electron microscopy experiments may contain atoms whose position can't be elucidated and appear as gaps in certain representations of the protein. Protein structures may have alternate conformations of side chains, which, when displayed in stick rendering, appear as two groups protruding off of the same amino acid backbone. Even short sections of backbone may have such alternative conformations, and sometimes ligands are superimposed in the active site in more than one binding conformation.
For a crystal structure, the deposited 3D coordinates include all components of the asymmetric unit, which provides enough information to reproduce the repeating unit of a protein crystal. Sometimes, this structure will contain additional protein chains compared to the biologically active form of the protein (e.g., fetal hemoglobin mutant, PDB ID: 4MQK). Conversely, some programs may not automatically load all chains of the biologically active unit. For example, the SARS-CoV2 main protease (PDB ID: 6Y2E) loads half of the biologically active dimer (made up of two protein chains) when fetched using the commands described in this protocol in ChimeraX, PyMOL, and Jmol. Though slight modification of the command will load the biologically active dimer, this consideration may not be straightforward for the novice modeling program user. A different issue that can arise is in the identification of the active site or substrate itself. Crystallographic experiments are carried out using a variety of molecules, which may be modeled into the final structure. For example, sulfate molecules may bind phosphate binding sites in the active site, or they may bind other regions that are not relevant to the mechanism. These molecules may obscure the correct identification of the active site itself and may even suggest to the student that they are part of the mechanism.
Presumably, the user will wish to apply this procedure to other active/binding sites. To apply this protocol in the future work involving the analysis of new protein active sites, the user will need to identify which of the bound ligands are relevant to function. Some ligands are not associated with protein function, and instead are a result of the solvent or crystallization conditions used to conduct the experiment (e.g., the potassium ion present in the 3FGU model). The key ligands should be identified by consulting the original manuscript. With practice and, where applicable, an understanding of the line command syntax, a user will be able to apply the protocol for the desired modeling program to any enzyme active site, and to model other macromolecules of their choice.
Identifying and analyzing bound substrates and ligands is central to the elucidation of molecular mechanisms and structure-based drug design efforts, which have directly led to improvements in treatments for disease, including acquired immunodeficiency syndrome (AIDS) and COVID-1947,48,49,50,51,52. While individual molecular visualization programs offer different interfaces and user experiences, most offer comparable features. It is important for the development of biomolecular visualization literacy that upper level biochemistry students become familiar with structure visualization and the tools to generate such images4,20,53. This allows students to move beyond the interpretation of two-dimensional images in textbooks and journal articles and to more easily develop their own hypotheses from structural data54, which will prepare developing scientists to address future public health issues and improve the understanding of biochemical processes.
In summary, this protocol details active site modeling using four leading free macromolecular modeling programs. Our community, BioMolViz, adopts a non-software-specific approach to biomolecular modeling. We specifically avoided a critique or comparison of program features, although a user sampling each program will likely find that they prefer certain aspects of macromolecular modeling in one program versus another. We invite readers to utilize the BioMolViz Framework, which details the biomolecular visualization-based learning goals and objectives targeted in this protocol, and explore resources for teaching and learning biomolecular visualization through the BioMolViz community website at http://biomolviz.org.
Disclosures
The authors declare that they have no relevant or material financial interests that relate to the research described in this paper.
Acknowledgements
Funding for this work has been provided by the National Science Foundation:
Improving Undergraduate STEM Education Grant (Award #1712268)
Research Coordination Networks in Undergraduate in Undergraduate Biology Education (Award # 1920270)
We are grateful to Karsten Theis, PhD, Westfield University, for helpful discussions about Jmol.
Materials
| Name | Company | Catalog Number | Comments |
| ChimeraX (Version 1.2.5) https://www.rbvi.ucsf.edu/chimerax/ | |||
| Computer | Any | ||
| iCn3D (web-based only: https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html) | |||
| Java (for Jmol) https://java.com/en/download/ | |||
| Jmol (Version 1.8.0_301) http://jmol.sourceforge.net/ | |||
| Mouse (optional) | Any | ||
| PyMOL (Version 2.4.1 - educational): https://pymol.org/2 educational use only version: https://pymol.org/edu/?q=educational |
References
- Loertscher, J., Green, D., Lewis, J. E., Lin, S., Minderhout, V. Identification of threshold concepts for biochemistry. CBE Life Sciences Education. 13 (3), 516-528 (2014).
- Jaswal, S. S., O’Hara, P. B., Williamson, P. L., Springer, A. L. Teaching structure: Student use of software tools for understanding macromolecular structure in an undergraduate biochemistry course: Teaching structure in undergraduate biochemistry. Biochemistry and Molecular Biology Education. 41 (5), 351-359 (2013).
- Tibell, L. A. E., Rundgren, C. -J. Educational challenges of molecular life science: Characteristics and implications for education and research. CBE Life Sciences Education. 9 (1), 25-33 (2010).
- Schönborn, K. J., Anderson, T. R. The importance of visual literacy in the education of biochemists. Biochemistry and Molecular Biology Education. 34 (2), 94-102 (2006).
- Anderson, T. R. Bridging the educational research-teaching practice gap: The importance of bridging the gap between science education research and its application in biochemistry teaching and learning: Barriers and strategies. Biochemistry and Molecular Biology Education. 35 (6), 465-470 (2007).
- Schönborn, K. J., Anderson, T. R. Bridging the educational research-teaching practice gap: Foundations for assessing and developing biochemistry students’ visual literacy. Biochemistry and Molecular Biology Education. 38 (5), 347-354 (2010).
- Bateman, R. C., Craig, P. A. Education corner: A proficiency rubric for biomacromolecular 3D literacy. PDB Newsletter. 45, 5-7 (2010).
- Mnguni, L., Schönborn, K., Anderson, T. Assessment of visualization skills in biochemistry students. South African Journal of Science. 112, 1-8 (2016).
- Craig, P. A., Michel, L. V., Bateman, R. C. A survey of educational uses of molecular visualization freeware. Biochemistry and Molecular Biology Education. 41 (3), 193-205 (2013).
- Loertscher, J., Villafañe, S. M., Lewis, J. E., Minderhout, V. Probing and improving student’s understanding of protein α-Helix structure using targeted assessment and classroom interventions in collaboration with a faculty community of practice. Biochemistry and Molecular Biology Education. 42 (3), 213-223 (2014).
- Abualia, M., et al. Connecting protein structure to intermolecular interactions: A computer modeling laboratory. Journal of Chemical Education. 93 (8), 1353-1363 (2016).
- Carvalho, I., Borges, A. D. L., Bernardes, L. S. C. Medicinal chemistry and molecular modeling: An integration to teach drug structure–activity relationship and the molecular basis of drug action. Journal of Chemical Education. 82 (4), 588(2005).
- Forbes-Lorman, R. M., et al. Physical models have gender-specific effects on student understanding of protein structure-function relationships. Biochemistry and Molecular Biology Education. 44 (4), 326-335 (2016).
- Terrell, C. R., Listenberger, L. L. Using molecular visualization to explore protein structure and function and enhance student facility with computational tools. Biochemistry and Molecular Biology Education. 45 (4), 318-328 (2017).
- Zhang, S., et al. Structure-based drug design of an inhibitor of the SARS-CoV-2 (COVID-19) main protease using free software: A tutorial for students and scientists. European Journal of Medicinal Chemistry. 113390, (2021).
- Roberts, J. R., Hagedorn, E., Dillenburg, P., Patrick, M., Herman, T. Physical models enhance molecular three-dimensional literacy in an introductory biochemistry course. Biochemistry and Molecular Biology Education. 33 (2), 105-110 (2005).
- Jenkinson, J., McGill, G. Visualizing protein interactions and dynamics: Evolving a visual language for molecular animation. CBE Life Sciences Education. 11 (1), 103-110 (2012).
- Bussey, T. J., Orgill, M. What do biochemistry students pay attention to in external representations of protein translation? The case of the Shine–Dalgarno sequence. Chemistry Education Research and Practice. 16 (4), 714-730 (2015).
- Harle, M., Towns, M. H. Students’ understanding of primary and secondary protein structure: Drawing secondary protein structure reveals student understanding better than simple recognition of structures. Biochemistry and Molecular Biology Education. 41 (6), 369-376 (2013).
- Dries, D. R., et al. An expanded framework for biomolecular visualization in the classroom: Learning goals and competencies. Biochemistry and Molecular Biology Education. 45 (1), 69-75 (2017).
- The BioMolViz Framework. BioMolViz. , Available from: http://biomolviz.org/framework (2021).
- Procko, K., et al. Meeting report: BioMolViz workshops for developing assessments of biomolecular visual literacy. Biochemistry and Molecular Biology Education. 49 (2), 278-286 (2021).
- Jmol: an open-source Java viewer for chemical structures. , Available from: http://www.jmol.org/ (2021).
- Wang, J., et al. iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics. 36 (1), 131-135 (2020).
- PyMOL . The PyMOL Molecular Graphics System. Version 2.0. , Schrödinger, LLC. (2021).
- Goddard, T. D., et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science. 27 (1), 14-25 (2018).
- Pettersen, E. F., et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Science. 30 (1), 70-82 (2021).
- Petit, P., et al. The active conformation of human glucokinase is not altered by allosteric activators. Acta Crystallographica. Section D. 67 (11), 929-935 (2011).
- Corey, R. B., Pauling, L. Molecular models of amino acids, peptides and proteins. Review of Scientific Instruments. 24, 621-627 (1953).
- Koltun, W. L. Precision space-filling atomic models. Biopolymers. 3 (6), 665-679 (1965).
- Hodis, E., et al. Proteopedia - a scientific 'wiki' bridging the rift between three-dimensional structure and function of biomacromolecules. Genome Biology. 9 (8), 1-10 (2008).
- Prilusky, J., et al. Proteopedia: A status report on the collaborative, 3D web-encyclopedia of proteins and other biomolecules. Journal of Structural Biology. 175 (2), 244-252 (2011).
- Martz, E. FirstGlance in Jmol. , Available from: https://www.bioinformatics.org/firstglance/fgij/ (2021).
- Jmol User Design Environment (JUDE). MSOE Centerfor BioMolecular Modeling. , Available from: https://cbm.msoe.edu/modelingResources/jmolUserDesignEnvironment/#forward (2021).
- Castro, C. R., et al. A practical guide to teaching with Proteopedia. Biochemistry and Molecular Biology Education. 49 (5), 707-719 (2021).
- Berman, H. M., et al. The protein data bank. Nucleic Acids Research. 28, 235-242 (2000).
- The Protein Data Bank. , Available from: https://www.rcsb.org/ (2021).
- Wang, Y., et al. MMDB: 3D structure data in Entrez. Nucleic Acids Research. 28 (1), 243-245 (2000).
- iCn3D Help Page. , Available from: https://www.ncbi.nlm.nih.gov/Structure/icn3d/docs/icn3d_help.html (2021).
- MSOE Center for BioMolecular Modeling Jmol Training Guide. , Available from: https://cbm.msoe.edu/modelingResources/jmolTrainingGuide/started.html (2021).
- The Online Macromolecular Museum. , Available from: http://earth.callutheran.edu/Academic_Programs/Departments/BioDev/omm/exhibits.htm (2021).
- Jmol/JSmol Interactive Scripting Documentation. , Available from: https://chemapps.stolaf.edu/jmol/docs/ (2021).
- PyMOL Wiki. , Available from: https://pymolwiki.org/index.php/Main_Page (2021).
- PyMOL Advanced Scripting Workshop by Schrödinger. , Available from: https://pymol.org/tutorials/scripting/index.html (2021).
- UCSF ChimeraX User Guide. , Available from: https://www.cgl.ucsf.edu/chimerax/docs/user/index.html (2021).
- UCSF ChimeraX Tutorials. , Available from: https://www.rbvi.ucsf.edu/chimerax/tutorials.html (2021).
- Kuntz, I. D. Structure-based strategies for drug design and discovery. Science. 257 (5073), 1078-1082 (1992).
- Structure-based drug discovery: an overview. Hubbard, R. E. , (2006).
- Patrick, G. L. An introduction to medicinal chemistry, 6th ed. , Oxford University Press. (2017).
- Van Montfort, R. L., Workman, P. Structure-based drug design: aiming for a perfect fit. Essays in Biochemistry. 61 (5), 431-437 (2017).
- Holdgate, G. A., Meek, T. D., Grimley, R. L. Mechanistic enzymology in drug discovery: a fresh perspective. Nature Reviews. Drug Discovery. 17 (2), 115-132 (2018).
- Wang, M. Y., et al. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Frontiers in Cellular and Infection Microbiology. 10, (2020).
- White, B., Kim, S., Sherman, K., Weber, N. Evaluation of molecular visualization software for teaching protein structure differing outcomes from lecture and lab: Differing outcomes from lecture and lab. Biochemistry and Molecular Biology Education. 30 (2), 130-136 (2002).
- Canning, D. R., Cox, J. R. Teaching the structural nature of biological molecules: Molecular visualization in the classroom and in the hands of students. Chemistry Education Research and Practice. 2 (2), 109-122 (2001).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved