Method Article
Surveying Low-Cost Methods to Measure Lifespan and Healthspan in Caenorhabditis elegans
In This Article
Summary
Caenorhabditis elegans serve as an excellent model system with robust and low-cost methods for surveying healthspan, lifespan, and resilience to stress.
Abstract
The discovery and development of Caenorhabditis elegans as a model organism was influential in biology, particularly in the field of aging. Many historical and contemporary studies have identified thousands of lifespan-altering paradigms, including genetic mutations, transgenic gene expression, and hormesis, a beneficial, low-grade exposure to stress. With its many advantages, including a short lifespan, easy and low-cost maintenance, and fully sequenced genome with homology to almost two-thirds of all human genes, C. elegans has quickly been adopted as an outstanding model for stress and aging biology. Here, several standardized methods are surveyed for measuring lifespan and healthspan that can be easily adapted into almost any research environment, especially those with limited equipment and funds. The incredible utility of C. elegans is featured, highlighting the capacity to perform powerful genetic analyses in aging biology without the necessity of extensive infrastructure. Finally, the limitations of each analysis and alternative approaches are discussed for consideration.
Introduction
Since the time of the publication of 'The genetics of Caenorhabditis elegans', one of the most influential articles by Sydney Brenner in 1974, this microscopic worm has been considered an outstanding model system to study biological mysteries1. In 1977, Michael R. Klass published the method for measuring the lifespan of C. elegans and established this model system to study aging2. The investigation to understand the relationship between stress and longevity has started with the identification of a single mutation in the age-1 gene, which resulted in a lifespan extension in C. elegans3. Furthermore, contemporary studies have identified other lifespan-increasing mutations, which revealed long-lived mutant worms that exhibit increased resistance to stress4,5,6. With its many advantages including a short lifespan, easy maintenance, completely sequenced genome containing homology to about two-thirds of all human disease-causing genes, availability and ease of using RNA interference (RNAi) libraries, and physiological similarity with humans7,8,9, C. elegans has quickly been adopted as an outstanding model for stress and aging biology.
Perhaps the greatest utilities of C. elegans are its extremely low cost of maintenance, ease of experimentation, and the variety of genetic tools available for studies. C. elegans are typically grown on a solid agar medium with an E. coli food source. Two commonly used E. coli strains are standard OP50, a B strain that is perhaps the most commonly used10, and HT115, a K-12 strain that is used primarily for RNAi experiments11,12. The HT115 K-12 strain carries a deletion in RNAIII RNase, a mutation that is essential for RNAi methods, where plasmids expressing dsRNA corresponding to individual C. elegans genes are used. The dsRNA feeding vectors allow for robust knockdown of C. elegans genes without the need for complex crosses or genome editing, as bacteria carrying these plasmids can be directly fed to nematodes. Thousands of these bacterial RNAi vectors exist in the HT115 background, including the most popular Vidal RNAi library with >19,000 individual RNAi constructs13 and the Ahringer RNAi library with 16,757 RNAi constructs14. However, the OP50 and HT115 bacterial diets have major differences in metabolic profile, including differences in Vitamin B1215,16. Therefore, it is recommended to perform all experiments on a single bacteria source, if possible, to avoid gene-diet interactions that may introduce multiple confounding factors as previously described17,18,19. Due to its ease, animals are maintained on OP50 for all the experimental conditions described here, but all experiments are performed on HT115 as previously described20. Briefly, animals are maintained at OP50 and transferred to HT115 post-synchronization (after bleaching) for consistency between RNAi vs. non-RNAi experiments. Alternatively, an RNAi-competent OP50 strain carrying a similar deletion of RNAIII RNase found in the E. coli K12 HT115 strain can also be used21.
Perhaps one major limitation to RNAi experiments in C. elegans is the concern of knockdown efficiency. While knockdown efficiency can be validated via qPCR or western blotting, these require expensive equipment and reagents and are limited to bulk analysis. This is even more of a concern looking at specific cells, such as neurons, which are refractory (less sensitive) to RNAi. While RNAi efficiency in specific cells can be enhanced via overexpression of SID-1, the transmembrane protein essential for dsRNA uptake22, this is still limited to the cell type-specific expression patterns of the promoters used for these constructs, and thus gene knockouts and mutations are the most foolproof means of depleting gene functions. Beyond RNAi-mediated knockdown, C. elegans are also highly amenable to genome editing with CRISPR-based strategies23,24,25 and transgenic construct overexpression through microinjections, with the option to integrate transgenic constructs through irradiation or transposon-based integration26,27,28,29. However, these methods require expensive microinjection equipment, and the high cost of guide RNAs or Cas9 enzyme may prohibit these methods in institutions with limited funding. Instead, thousands of transgenic lines and mutants are readily available for a few dollars both at the Caenorhabditis Genetics Center (CGC) and National Bioresource Project (NBRP). The NBRP offers isolated mutants for a large number of C. elegans genes, including published and therefore verified mutant strains, mutants derived from pilot projects, and mutants that have yet to be characterized. In contrast, CGC is a depository of mostly published and established C. elegans lines from the research community. Both of them ship strains worldwide at very reasonable rates and offer a wide variety of options for those with limited capacity to synthesize strains in-house.
Here, a curated methods collection is offered, which are likely to be the lowest cost methods for assaying lifespan and healthspan in C. elegans. All the methods presented here require low-cost equipment and supplies, and only utilize strains readily available from the CGC. Perhaps most prohibitive for longevity and survival assays in C. elegans is the cost of Nematode Growth Media (NGM) plates. Since C. elegans are hermaphrodites and self-fertilize, standard survival assays require that adult animals be continuously moved away from their progeny to avoid contamination from offspring. Not only is this process time-consuming, it can become expensive due to the necessity of approximately 100 plates per condition to run a single lifespan assay. Here, two alternatives are provided: utilization of the temperature-sensitive germline-less mutant, glp-4(bn2), or chemical sterilization using 5-fluoro-2'-deoxyuridine (FUDR). glp-4 encodes a valyl aminoacyl tRNA synthetase, and the temperature-sensitive glp-4(bn2) are reproductively deficient at restrictive temperatures due to decreased protein translation30,31. FUDR is a robust method to chemically sterilize C. elegans by preventing DNA replication, thus inhibiting reproduction32. Although FUDR can be prohibitively expensive for some labs, only a small amount is required to chemically sterilize worms, and its stability in powder form may make it feasible for most groups. Utilizing the temperature-sensitive glp-4(bn2) mutant is certainly the cheapest option, as the only requirement is an incubator to shift the animals to the restrictive 25 °C; however, it should be noted that growth at 25 °C may cause mild heat-stress33,34. Regardless of the method, using sterile animals can significantly decrease the costs of consumables required for age-related assays.
To study aging, standard lifespan assays are conventional as paradigms that alter longevity have direct impacts on aging. However, measurements of healthspan and stress tolerance present additional information on the health of the organism. Here, several methods are offered to measure healthspan: 1) fecundity as a measure of reproductive health; 2) brood size as a measure of developmental health and viability of laid offspring; and 3) locomotory behavior as a measure of muscle function and motility, both of which are directly correlated with aging. Additionally, assays of stress tolerance are offered: survival to ER stress, mitochondrial/oxidative stress, and thermal stress survival. Indeed, animals with increased resistance to ER stress35,36, mitochondrial stress37, and thermal stress38 exhibit increased lifespan. ER stress is applied by exposing C. elegans to tunicamycin, which blocks N-linked glycosylation and causes the accumulation of misfolded proteins39. Mitochondrial/oxidative stress is induced by exposure to paraquat, which induces superoxide formation specifically in the mitochondria40. Heat stress is applied through the incubation of animals at 34-37 °C33,41. All the assays described here can be performed with minimal equipment and funds, and offer a variety of tools to study aging in diverse groups.
Protocol
1. Growth and maintenance of C. elegans
- Pouring Nematode Growth Media (NGM) plates
- Grow C. elegans on standard 2% agar plates with Nematode Growth Media (NGM) consisting of 1 mM CaCl2, 5 µg/mL cholesterol, 25 mM KPO4 (pH 6.0), 1 mM MgSO4, 0.25% w/v Peptone, and 51.3 mM NaCl.
- For 1 L of NGM-agar plates, measure out 2.5 g of Peptone, 3.0 g of NaCl, and 20 g of agar into a 1 L flask with a stir bar.
NOTE: It is recommended to standardize a specific agar source, as we have seen variability in stiffness between brands, which may affect reproducibility. Here, Bacto-Agar is strictly used. Additionally, it is recommended to add agar directly into the flask being autoclaved, as agar will not dissolve completely without heating, and transferring solution containing agar will result in loss of agar and errors in concentration. - Add dH2O up to 970 mL.
NOTE: 30 mL of liquid additives post-sterilization will bring the final volume to 1 L. In our hands, ~951 mL of dH2O will be required to reach 970 mL of the final volume. - Sterilize NGM-agar solution using a standard autoclave or media sterilizer for efficient sterilization.
NOTE: At this point, sterile NGM-agar can be stored for several months at room temperature. If stored, NGM-agar can be reliquefied in a microwave with 15-45 s pulses to prevent the solution from boiling over, or in a heated water bath. - Let the solution stir until it is cooled to 60-75 °C. Stirring while cooling is important to prevent uneven cooling, which may cause some agar to solidify.
- While the solution cools, heat a water bath or bead bath to 65-70 °C.
- Once the solution cools to 60-75 °C, add liquid additives: 2.0 mL of 0.5 M CaCl2, 1 mL of 5 mg/mL cholesterol, 25 mL of 1 M KPO4 (pH 6.0), and 0.5 M MgSO4 (see Table 1 for recipes for all reagents), and allow solution to mix for ~5 min to ensure complete mixing.
NOTE: Drugs can also be included in plates here (e.g., add 1 mL of 100 mg/mL carbenicillin and 1 mL of 1 M IPTG; add 10 mL of 2.5 mg/mL tunicamycin; add 10 mL of 400 mM paraquat). - Submerge flask containing NGM-agar in a 65-70 °C water bath to prevent NGM-agar from solidifying while pouring into plates.
- Pipette 9-11 mL of solution into each 60 mm plate, or 20-30 mL of solution into each 100 mm plate.
NOTE: It is recommended to use the smallest pipette volume available to avoid NGM leaks caused by air expanding in the pipette. Pipetting up 1-2 mL more media than will be added to each plate to avoid completely emptying the pipette will help prevent bubble formation. Alternatively, plates can be hand-poured directly from the bottle into a plate, but pipetting is highly recommended to ensure plates with equal volume. Equal volume plates are important to ensure similar concentrations of solutions when using methods where solutions are applied directly on top of a plate (see step 1.1.17). Equal volumes also allow for easy microscopy to maintain a similar focal plane across plates. - Place the pipette back in the heated solution to maintain the temperature and to prevent NGM-agar from solidifying.
- Repeat the above two steps for all the plates.
- Allow NGM-agar plates to solidify overnight.
- After plates have solidified, store plates for up to 3 months at 4 °C or move on to step 1.1.14 for seeding plates with bacteria. Store plates in sealed containers to help retain moisture to maintain plate quality.
- Grow a culture of OP50 in lysogeny broth (LB) or equivalent media of choice for 24-48 h at ambient temperature (~22-25 °C) or grow a culture of HT115 in LB + antibiotics (ampicillin/carb + tetracycline is recommended for HT115) with shaking at 37 °C for 12-16 h.
NOTE: It is recommended to grow OP50 at room temperature because a more aggressive growth of OP50 has been found at 37 °C, which affects C. elegans lifespan. In contrast, HT115 has a slower growth rate and makes less dense cultures; thus, it is recommended to grow HT115 at 37 °C with shaking. - Seed a volume of 100-200 µL of a saturated OP50/HT115 culture onto a 60 mm plate, or 1 mL for a 100 mm plate.
- Let plates dry overnight on a benchtop and allow for an additional day to dry if plates are still wet. Store plates in sealed containers at 4 °C for ~2 months.
- Optional: Add drugs directly onto seeded NGM-agar plates (e.g., 100 µL of 10 mg/mL FUDR solution) to chemically sterilize worms.
- Maintaining C. elegans stocks
- Properly label the bottom of a seeded NGM-agar plate. Label the edges on the bottom of the plate to prevent obstructing the passage of light on standard dissection microscopes.
- Using a standard dissection microscope, scoop bacteria of choice onto C. elegans pick.
NOTE: In this protocol, a pick comprised of a 90%/10% platinum/iridium wire attached to the end of a glass Pasteur pipette was utilized. - Using the bacteria, collect 10-20 eggs, L1, L2, or L3 animals, and transfer them onto a newly labeled seeded NGM-agar plate.
NOTE: It is best to collect younger animals; in the authors' experience, for standard wild-type animals, moving 10-20 eggs/young animal will allow the plate to grow at 15 °C without starvation. For transgenic or mutant animals with decreased fecundity, more animals should be moved into the plate. - For animals with wild-type fecundity and grown at 15 °C, repeat steps 1.2.1-1.2.3 every 7 days to maintain a weekly stock. For animals grown at 20 °C, repeat steps 1.2.1-1.2.3 every 4-5 days to prevent starvation.
- Synchronizing worms via bleaching
NOTE: A full, 60 mm NGM-agar plate (e.g., 1-week-old stock plates grown at 15 °C) will provide a sufficient number of animals for most standard assays described. Generally, one gravid adult (adult full of eggs) will provide 10-15 eggs42, and a full, 60 mm NGM-agar plate has anywhere from 100-200 gravid adults, providing ~1000-2000 eggs.- For larger-scale experiments requiring more animals, cut a full 60 mm NGM-agar into four to six equal pieces and chunk them onto seeded 100 mm plates for expansion.
NOTE: Here, chunking refers to cutting a piece of the NGM-agar plate containing worms and moving the entire chunk of agar + worms onto a new plate, worm-side down to allow the worms to crawl onto the new plate. As a frame of reference, animals with wild-type fecundity will produce a full 100 mm plate if grown at 20 °C for 2-3 days after chunking. - To begin collecting the nematodes, pour a small amount of M9 solution (Table 1) onto plates containing worms, taking care not to overfill the Petri dish. Swirl the M9 solution gently to loosen worms off bacterial lawns.
- Collect gravid adult worms with a serological pipette, taking care not to pierce the agar with the pipette tip.
NOTE: Glass serological pipettes are recommended, as C. elegans tend to stick to plastic. If glass pipettes are not available, it is recommended to start with a larger number of animals than needed, as some will be lost due to sticking to plastic pipettes. - Pellet the animals by centrifugation for 30 s at 1,100 x g. Aspirate the supernatant.
NOTE: The C. elegans pellet is very loose, so take care not to shake or disrupt the pellet while aspirating the supernatant. - While animals are centrifuging, prepare 5 mL of bleaching solution per strain (see Table 1 for recipe details); for 5 mL of solution, mix 1.5 mL of 6% sodium hypochlorite (bleach), 0.75 mL of 5 M NaOH or KOH, and 2.75 mL of dH2O.
CAUTION: Sodium hypochlorite and high concentration hydroxide solutions are corrosive, and thus it is recommended to wear gloves and a lab coat when handling. - Add 5 mL of bleaching solution to the worm pellet/M9 mixture.
- Check the worms under a dissecting microscope every few minutes until all the adult worm bodies have been dissolved and only eggs are left in the mix. Shake the worm/bleach mix vigorously to speed up the bleaching process.
NOTE: Leaving eggs inside a bleach mix for extended periods will result in damaging the eggs and will affect the viability of the animals. For wild-type animals, bleaching usually takes 4-6 min with shaking. Thus, it is recommended to check the animals under a microscope at 30 s intervals starting from the 4 min mark. - Pellet the eggs by spinning down the egg/bleach mix for 30 s at 1,100 x g.
NOTE: Some 15 mL conical tubes have gradient lines on the inside of the tube. For these tubes, it is recommended to spin eggs at a higher speed (e.g., 30 s at 2,000 x g) to ensure that eggs pellet to the bottom of the tube and do not remain on gradient lines. - Aspirate out the bleaching solution.
NOTE: An egg pellet is stiffer than a worm pellet, but can still be disrupted easily. So, take care not to shake the tube after centrifuging. - Wash eggs by adding M9 solution up to 15 mL and inverting the tube four or five times to ensure that the eggs are fully dispersed in the M9 solution.
- Pellet eggs by centrifuging for 30 s at 1,100 x g and aspirate out M9 solution.
- Repeat the above two steps for a total of four washes to eliminate any bleach from the egg mix.
- Resuspend the eggs in 100 µL to 2 mL of M9 solution (i.e., depending on the total number of worms bleached) after the final wash. Shake eggs thoroughly to break up clumps and ensure that the pellet is fully resuspended.
- Alternatively, animals can be L1 arrested for a tighter temporal synchronization; for L1 arresting, add M9 solution to the egg pellet to ~10 mL in a 15 mL conical tube. Let the worms spin in a rotator for up to 24 h at 20 °C or ambient temperature. L1 animals generally take half a day less to reach adulthood compared to the timing of eggs described in step 1.3.16.
- Approximate the egg concentration (or L1 concentration; see step 1.3.14) by pipetting 4 µL of egg/M9 mixture onto an NGM-plate seeded with bacteria. Count and calculate how many eggs are present per µL plated. Repeat the counting three or four times to improve the approximation.
NOTE: Approximating the egg concentration will ensure that enough animals are plated for appropriate sample size for experiments without overplating, which will cause starvation. - Based on the approximation, plate the appropriate number of eggs onto NGM-agar plates seeded with bacteria of choice. For OP50 plates, plate a maximum of 200 animals on a 60 mm plate and 1,000 animals on a 100 mm plate. For HT115 plates, plate a maximum of 150 animals on a 60 mm plate and 600 animals on a 100 mm plate.
NOTE: These are approximated numbers based on our lab conditions, and numbers may change based on thickness of bacterial lawn. Eggs grown at 15 °C will take ~5 days to reach day 1 adulthood (~140 h to reach egg-laying maximal, gravid adult stage). Eggs grown at 20 °C will take ~4 days to reach day 1 adulthood (~96 h to reach egg-laying maximal, gravid adult stage). Eggs grown at 25 °C will take ~3.5 days to reach day 1 adulthood (~62 h to reach egg-laying maximal, gravid adult stage).
- For larger-scale experiments requiring more animals, cut a full 60 mm NGM-agar into four to six equal pieces and chunk them onto seeded 100 mm plates for expansion.
- Egg-lay as an alternative method to synchronize C. elegans populations
- If bleaching protocols are not feasible (e.g., no centrifuge available), as an alternative method to synchronize populations of C. elegans, perform an egg-lay procedure. Keep in mind that this protocol is more labor-intensive and will result in smaller yields of animals.
- For egg-laying, place 8-12 gravid adults onto a standard NGM-agar plate seeded with bacteria of choice and document the exact number of animals placed onto a plate.
NOTE: Egg-lay procedures should be performed at the temperature that will be used for experimentation. - Allow animals to lay eggs for 4-8 h.
NOTE: The duration animals are left on the plate can be adjusted when needed. For example, a larger number of animals can be put on a plate for a shorter egg-lay duration when less time is available. C. elegans generally lay eggs in bursts, which can be estimated at a rate of approximately five eggs/h for animals with wild-type fecundity43. Follow recommendations in step 1.3.16 to avoid overplating animals. - Remove all adult animals from the plate.
NOTE: Any adult animals left on the plate will continue to lay eggs, resulting in an unsynchronized population. - Place eggs at 15 °C for ~5 days or 20 °C for ~4 days to reach day 1 adulthood.
2. Measuring longevity in C. elegans
- Standard lifespan
- Prepare NGM-agar plates by seeding plates with 100 µL of bacteria of choice. For consistency, ensure that the same bacteria is used across all replicates. Since worms are moved every day during the egg-laying stages of adulthood, seed five to seven sets of NGM-agar plates for the duration of the lifespan, and two to four plates per strain to grow animals to adulthood (i.e., if using eight plates of 15 animals for lifespans, one needs to seed 40-56 plates per condition).
- Allow plates to dry overnight before storage.
NOTE: It is recommended that the plates be stored at 4 °C, and the required number of plates be removed from cold storage daily to prevent bacteria from making thick lawns that can make moving/counting lifespans difficult. Ensure that plates are warmed up prior to plating worms. - Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4.
- Move 10-15 day 1 adult animals onto 8-12 plates each. For a standard lifespan, start with ~120 animals to ensure that sample size does not drop too far below 100 after censorship events (e.g., eight plates of 15 animals = 120 animals; 12 plates of 10 animals = 120 animals).
NOTE: In the current case, 10-15 animals are a manageable number for most investigators, although six plates of 20 animals is also feasible to decrease the cost of consumables. - For the first 7-8 days or until progeny are no longer visible, move adult animals away from their progeny every 1-2 days.
NOTE: Animals can be moved every other day to save materials, but care must be taken to ensure that eggs/larval animals are not transferred with the adult to prevent contamination of adult populations with progeny. In this study, the simplest method is to move animals every day from days 1-3 when egg-laying is at its maximal, and then switch to moving animals every other day for days 5-8 when egg-laying is minimal. With this method, it is not imperative to prevent transfer of eggs/larval animals during days 1-3 since the adults will be moved every day, and eggs/larvae cannot develop to adulthood in 1 day. - After animals have stopped producing progeny, score the lifespans every other day until all animals have been scored as dead or censored. Remove all dead or censored animals from the plate to avoid confusion and recounting the same animal.
NOTE: Death is scored as animals that exhibit no movement when gently touched with a pick. Censorship is scored as animals that are bagged, exhibit vulval/intestinal protrusions, or crawled up to the sides of the plate where they desiccate.
- Lifespans with chemical sterilization using FUDR
- Prepare NGM-agar plates by seeding plates with 100 µL of bacteria of choice. For consistency, ensure that the same bacteria is used across all replicates. Seed 8-12 plates per strain for lifespan experiments, and two to four plates per strain to grow animals to adulthood. Allow plates to dry overnight.
- Add 100 µL of 10 mg/mL FUDR onto the middle of bacterial lawn for the 8-12 plates that will be used for lifespan assay. Remember to leave two to four plates without FUDR as starter plates to allow animals to grow to adulthood. Let plates dry overnight.
CAUTION: FUDR blocks DNA synthesis, and thus it is recommended to wear gloves when handling. - Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4.
NOTE: These animals need to be grown on plates without FUDR, as FUDR will cause animals to arrest/die. - Move 10-15 day 1 adult animals onto 8-12 plates each, containing FUDR. For a standard lifespan, start with ~120 animals to ensure that sample size does not drop too far below 100 after censorship events (e.g., eight plates of 15 animals = 120 animals; 12 plates of 10 animals = 120 animals).
NOTE: Animals can also be moved onto FUDR at the L4 stage if it is imperative that progeny formation be completely avoided, but animals must not be moved too early as this will cause animals to be at higher risk for vulval/intestinal protrusions and will increase censorship. - Score lifespans every other day until all animals have been scored as dead or censored. Remove all dead or censored animals from the plate to avoid confusion and recounting the same animal.
NOTE: For FUDR lifespans, any progeny can be ignored as they will arrest at the L1 stage and eventually die.
- Lifespans using temperature sensitive sterile mutants
- Prepare NGM-agar plates by seeding plates with 100 µL of bacteria of choice. For consistency, ensure that the same bacteria is used across all replicates. Seed 8-12 plates per strain for lifespan experiments, and 2-4 plates per strain to grow animals to adulthood. Let plates dry overnight.
- Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4. Remember to grow animals at the restrictive temperature of 25 °C to ensure that animals are sterile.
- Move 10-15 day 1 adult animals onto 8-12 plates each. For a standard lifespan, start with ~120 animals to ensure that sample size does not drop too far below 100 after censorship events (e.g., eight plates of 15 animals = 120 animals; 12 plates of 10 animals = 120 animals).
- Score lifespans every other day until all animals have been scored as dead or censored. Remove all dead or censored animals from the plate to avoid confusion and recounting the same animal.
NOTE: When dealing with short-lived strains, it is recommended to score lifespans every day as lifespans at 25 °C are much shorter and thus the dynamic range is limited. In authors' experience, animals can be shifted back to 20 °C after day 2, and animals will remain sterile if it is preferable to score lifespans at 20 °C.
3. Measuring healthspan in C. elegans
- Measurements of locomotory behavior via thrashing
- Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4.
- Move a small colony of day 1 adult worms onto an NGM-agar plate under a dissecting scope onto 10-20 µL of M9 solution. 10-15 animals are recommended as a manageable number of animals to count.
- Focusing on one worm at a time, count the number of times the specimen switches from a concave to convex formation in 15 s. Use a hand counter and a timer so focus can be placed on the worm for the duration of the assay.
NOTE: A video of the plate may be recorded for more thorough/easier analysis. For example, standard microscope eyepiece attachments are available for most smartphones and digital cameras ($15-$30), and these are a great option to video thrashing at a low cost. - Repeat step 3.1.3 for the other worms in the liquid, averaging out a total motility rate for 10-15 worms. For higher sample size, repeat steps 3.1.2-3.1.4.
- Age out worms to desired age. Similar methods for lifespan assays described in steps 2.1-2.3 can be used for aging out worms. Repeat steps 3.1.2-3.1.4 to assay thrashing at desired ages.
NOTE: An alternative method for step 3.1.2 is to add ~30 µL or more of M9 solution onto a group of worms on a plate. This will save time from having to manually transfer worms, although due to random chance of where the worms are on a single plate, there is no guarantee that a group of worms will remain at a single point on the plate.
- Measurements of fecundity (egg count) in C. elegans
- Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4. Assays for egg count start at the L4 stage, which is ~1 day prior to day 1 adulthood (~3 days at 15 °C or ~2 days at 20 °C after L1 arresting).
- Single out L4 worms onto separate NGM-agar plates seeded with bacteria of choice. It is recommended that ~10-15 animals be used for a fecundity assay.
NOTE: It is recommended to dilute the bacteria of choice by 50% (i.e., not a saturated culture) to improve the egg visibility in the bacterial lawn. - Allow animals to grow overnight at 20 °C. Ensure that a newly seeded batch of plates is ready for the next day.
- On day 1 of adulthood, transfer adult worms onto fresh NGM-agar plates seeded with the diluted bacteria of choice.
NOTE: It is recommended to use freshly seeded plates, or to store plates at 4 °C until use to prevent thick bacterial lawns. - Count the total number of eggs laid on each NGM-agar plate.
NOTE: To aid in scanning the plate, a grid can be drawn on the lid of a plate and placed under the plate being scored for eggs. The plate can then be scanned along the grid lines to maintain orientation as the plate is moved and prevent recounting of any eggs. - Repeat steps 3.2.4-3.2.5 for 7-8 days or until eggs are no longer visible on the plate.
NOTE: For days 1-3 when egg-laying rates are high, it is recommended to move animals at least every 12 h and assay egg counts twice a day. However, this increases the amount of work and costs of consumables, and thus moving animals and measurements can be limited to once per day, but care must be taken to ensure all eggs and hatched animals are counted properly. Any hatched animals are counted as eggs for the purpose of this assay.
- Measurement of brood size (development) of C. elegans progeny
- Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4. Assays for brood size start at the L4 stage, which is ~1 day prior to day 1 adulthood (~3 days at 15 °C or ~2 days at 20 °C after L1 arresting).
- Single out L4 worms onto separate NGM-agar plates seeded with bacteria of choice. It is recommended that ~10-15 animals be used for a fecundity assay.
- Allow animals to grow overnight at 20 °C. Ensure that a newly seeded batch of plates is ready for the next day.
- On day 1 of adulthood, transfer adult worms onto fresh NGM-agar plates seeded with bacteria of choice.
- Every 12-24 h (2x a day or 1x a day), transfer adult worms onto fresh NGM-agar plates seeded with bacteria of choice for 7-8 days or until progeny are no longer visible. Keep all the plates containing eggs at 20 °C.
- Repeat step 3.3.5 for 7-8 days or until progeny are no longer visible. Keep all the plates containing eggs at 20 °C.
NOTE: Progeny plates may also be stored at 15 °C to extend the time before they need to be scored. - Two days after transferring worms, count the developed progeny on the plates. Count developing worms at the L4 stage (i.e., 2 days after hatch at 20 °C) or earlier to ensure that the F2 generation (i.e., progeny of progeny) does not confound results. Count all worms that are alive.
- Remove all worms from the plate as they are counted. Maintain the plates for an additional 1-2 days before scoring them again to ensure that any animals with delayed hatching/development are not missed.
- Repeat step 3.3.7 for every egg-lay plate collected.
NOTE: The brood size assays can be conducted in conjunction with egg count assay (step 3.2) to minimize labor and costs of consumables by collecting two sets of data from one experiment. This will also allow for direct comparison of brood size and egg count within the same animals.
4. Measuring stress resilience in C. elegans
- Measurements of ER stress sensitivity using tunicamycin
- Prepare NGM-agar plates by seeding plates containing tunicamycin (see step 1.1.7, NOTE) with 100 µL of bacteria of choice.
CAUTION: Gloves should be worn when handling tunicamycin. - For consistency, ensure that the same bacteria is used across all replicates. Seed 8-12 tunicamycin plates per strain for survival assays, and two to four plates without tunicamycin per strain to grow animals to adulthood. Let plates dry overnight.
- Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4.
NOTE: Animals must be grown on plates without tunicamycin until day 1 of adulthood, as animals will arrest/die on tunicamycin. - Move 10-15 day 1 adult animals onto 8-12 plates each. For a standard survival assays, start with ~120 animals to ensure that sample size does not drop too far below 100 after censorship events (e.g., eight plates of 15 animals = 120 animals; 12 plates of 10 animals = 120 animals).
NOTE: Similar to FUDR assays, tunicamycin survival assays can be performed without moving animals, as tunicamycin causes death/arrest of L1 animals. However, when performing a DMSO control, progeny will develop on DMSO plates, so animals need to be moved daily or a sterilization technique will be required (identical methods used in section 2 for lifespans can be used for survival assays). - Survival assays are scored similar to lifespans. Remove all dead or censored animals from the plate to avoid confusion and recounting the same animal.
NOTE: Although it is possible to score animals every other day, since death occurs rapidly on tunicamycin, it is recommended to score survival assays daily.
- Prepare NGM-agar plates by seeding plates containing tunicamycin (see step 1.1.7, NOTE) with 100 µL of bacteria of choice.
- Measurements of mitochondrial/oxidative stress sensitivity using paraquat
- Prepare NGM-agar plates by seeding plates containing paraquat (see step 1.1.7; NOTE) with 100 µL of bacteria of choice.
CAUTION: Gloves should be worn when handling paraquat as it is an environmental hazard. Check with institution's environmental health and safety for requirements of discarding, as many research institutions will require specific discarding instructions for environmental hazards. - For consistency, ensure that the same bacteria is used across all replicates. Seed 8-12 plates per strain for survival assays, and two to four plates without paraquat per strain to grow animals to adulthood. Allow plates to dry overnight.
- Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4.
NOTE: Remember to grow animals on plates without paraquat until day 1 of adulthood; however, it is necessary to perform a sterilization technique or move adults away from progeny, as some animals can develop to adulthood on paraquat plates (see steps 2.2-2.3). - Move 10-15 day 1 adult animals onto 8-12 plates each. For a standard survival assay, start with ~120 animals to ensure that sample size does not drop too far below 100 after censorship events (e.g., eight plates of 15 animals = 120 animals; 12 plates of 10 animals = 120 animals).
- Survival assays are scored similar to lifespans. Remove all dead or censored animals from the plate to avoid confusion and recounting the same animal.
NOTE: Although it is possible to score animals every other day, since death occurs rapidly on paraquat, it is recommended to score survival assays daily. This is especially true when using glp-4(bn2) animals grown at 25 °C as death will occur very rapidly.
- Prepare NGM-agar plates by seeding plates containing paraquat (see step 1.1.7; NOTE) with 100 µL of bacteria of choice.
- Measurements of heat stress sensitivity (thermotolerance) using elevated temperatures
- Collect a synchronized population of C. elegans using a standard bleaching assay as described in steps 1.3 and 1.4.
- Pre-warm NGM-agar plates to 37 °C prior to moving animals onto plates by placing plates into a 37 °C incubator for at least 1 h.
- Move 10-15 day 1 adult animals onto four to six pre-warmed plates each. For a standard thermotolerance, start with ~60 animals (e.g., four plates of 15 animals = 60 animals; six plates of 10 animals = 60 animals)
- Place animals into a 37 °C incubator and score for death every 2 h. Death is defined as animals that exhibit no movement when gently touched with a pick. Remove all dead or censored animals from the plate to avoid confusion and recounting the same animal.
- Ensure that plates are removed from the 37 °C incubator for the minimal amount of time possible, as plates that are left at ambient temperature for long durations while scoring will alter thermotolerance results.
NOTE: It is recommended to pull out only one strain at a time to score, as the temperature of the agar should not change dramatically in the time it takes to score one strain. - Median thermotolerance is generally accomplished in 7-9 h; so, ensure a proper assay at 7 h, 9 h, and 11 h.
NOTE: While 1-5 h can be skipped, due to variability of incubators, the thickness of plates, and other confounding factors in each lab, it is important that timing is titrated carefully in each lab if timepoints are planned to be skipped. See reference 44 for a full guide on thermotolerance. - Alternatively, perform the thermotolerance assay at 34 °C instead of 37 °C.
NOTE: Median thermotolerance at 34 °C occurs much later (10-14 h in this study), which allows for thermotolerance assays to be prepared late at night (placed in a 34 °C incubator), and for scoring to begin early the next day. This allows for ~8 h of continued scoring rather than the typical 12 h period required for a 37 °C thermotolerance assay.
Results
C. elegans are an excellent model organism for aging research due to a large majority of aging mechanisms being conserved with humans. Importantly, they have a very low cost in maintenance and experimentation with minimal requirements for equipment and consumables, making them a coveted model system for institutions with limited funds. Moreover, a plethora of simple assays with shallow learning curves makes them an excellent system for even the youngest investigator with little to no experience. All these factors combined with the powerful genetics of C. elegans including the ease of genome editing, thousands of available mutants and transgenic animals at nominal costs, and available RNAi libraries for genetic knockdown of virtually every gene make them an ideal system for undergraduate institutions. Here, some of the lowest cost methods to study aging in C. elegans are surveyed, focusing primarily on assays with minimal equipment and consumable cost, as well as shallow learning curves. In fact, the entirety of the protocols and data collection were written/performed by junior investigators with <5 months of research experience, mostly undergraduate students.
Longevity studies in C. elegans are very simple due to the short lifespan of animals, ranging from 14-20 days. Importantly, lifespan assays are highly standardized and only require an incubator, a standard dissection microscope, a standard worm pick, and consumables for preparing NGM-agar plates. Perhaps the most cost-prohibitive aspect of lifespan measurements in C. elegans is consumables required. This is because C. elegans are hermaphrodites that self-fertilize; therefore, adults being tracked for longevity assays need to be moved away from progeny daily. However, animals can be sterilized by exposing them to FUDR or using mutants, such as the temperature-sensitive germline-less glp-4(bn2) mutant grown at the prohibitive 25 °C to reduce the amount of consumables required30,31,32. Here, lifespan assays were performed with FUDR or with the glp-4(bn2) germline-less mutants, which show similar results to standard lifespans performed on non-sterile animals. While the wild-type lifespans are not identical due to the effects of FUDR45 or growth at 25 °C on lifespan2, the short-lived hsf-1 knockdown animal reliably shows a significant decrease in lifespan for all conditions (Figure 1). hsf-1 encodes the heat-shock factor-1 transcription factor, which is involved in the regulation of the thermal stress response, and its knockdown results in a significant decrease in lifespan38,46.
While longevity is an important factor to consider in aging biology, often, longevity does not correlate with increased health, even in C. elegans47. Thus, as a complementary approach, we offer several methods of measuring organism health, including reproductive health, locomotory behavior, and stress resilience. Reproductive health can be measured in one of two ways. First, measurements of egg count will give a direct measurement of how many eggs are laid by a single self-fertilizing hermaphrodite. However, since animals produce more oocytes than sperm, some unfertilized eggs that would never produce viable progeny are also laid48. Therefore, to get a better understanding of the true reproductive capacity of an animal, measurements of the brood size provide a measure of how many viable offspring are produced. Oftentimes, increased stress resilience can actually decrease reproductive capacity, potentially due to the inherent effect of perceived stress on reproduction49. Similarly, a significant decrease in both the number of eggs laid and brood size is found in hsf-1 overexpression animals compared to wild-type controls (Figure 2A,B). In fact, some hsf-1 overexpression animals exhibit full sterility, providing evidence that reproductive health can be inversely correlated with longevity.
While reproductive health is important to understanding germline health, functional meiosis, and reproductive capacity, in general, there is no direct correlation between longevity and brood size50. Thus, as a complementary approach, locomotory behavior is offered as a gold-standard method for assaying C. elegans healthspan during aging51. There are many methods to measure locomotory behavior, but most methods require sophisticated cameras, tracking software, or expensive chemicals. In contrast, thrashing assays require virtually no equipment beyond what a standard C. elegans lab is equipped with: dissecting microscope, worm pick, pipette, and consumables for making NGM-agar plates. Thrashing rates provide a reliable method for measuring healthspan during aging, as measured by a significant decrease in thrashing in old animals compared to young animals (Figure 2C).
Finally, survival to stress assays is an additional physiological measurement of resilience. The capacity to activate stress responses generally declines during the aging process, making animals less resilient and more sensitive to stress. Thus, stress resilience can be used as a reliable proxy for organismal health. Here, methods are offered for surveying sensitivity to 1) ER stress in response to tunicamycin exposure, a chemical agent that blocks N-linked glycosylation and results in accumulation of misfolded proteins in the ER; 2) mitochondrial/oxidative stress through exposure to paraquat, a chemical agent that induces superoxide formation in mitochondria; and 3) thermal stress through exposure to elevated temperatures. For tunicamycin and paraquat assays, the drug is incorporated into the NGM-agar plate during plate production. For high concentrations of tunicamycin, progeny generally does not develop, and thus sterilization techniques do not need to be used. The protocol presented here recommends 25 ng/µL as a final concentration of tunicamycin, but for those with limited funds, 10 ng/µL also shows a significant reduction in survival (Figure 3A). Both concentrations limit progeny development, and thus no sterilization methods are needed, although the DMSO control will require a sterilization technique or movement of animals onto new plates. This is because tunicamycin toxicity prevents the development of progeny, but DMSO is virtually nontoxic, which allows progeny to develop fully when grown on tunicamycin.
For paraquat assays, either a sterilization technique or movement of animals is required as paraquat treatment does not prevent progeny from developing to adulthood. High levels of paraquat (4 mM) significantly shorten lifespan, while low levels of paraquat (0.25 mM) increase lifespan due to a hormetic effect (Figure 3B), consistent with previously published results52. Finally, thermotolerance assays only require an incubator that can reach 30-37 °C, and no additional reagents are required. Overexpression of hsf-1 increases thermotolerance at 37 °C (Figure 3C) as previously published53. However, as others have shown previously and from the present data, the major problem with thermotolerance assays is their variability. Many factors can contribute to variability within thermotolerance assays, including differences between incubators and the time animals spend outside of the incubator while scoring thermotolerance each hour. For a thorough guideline of thermotolerance, refer to the reference 41.
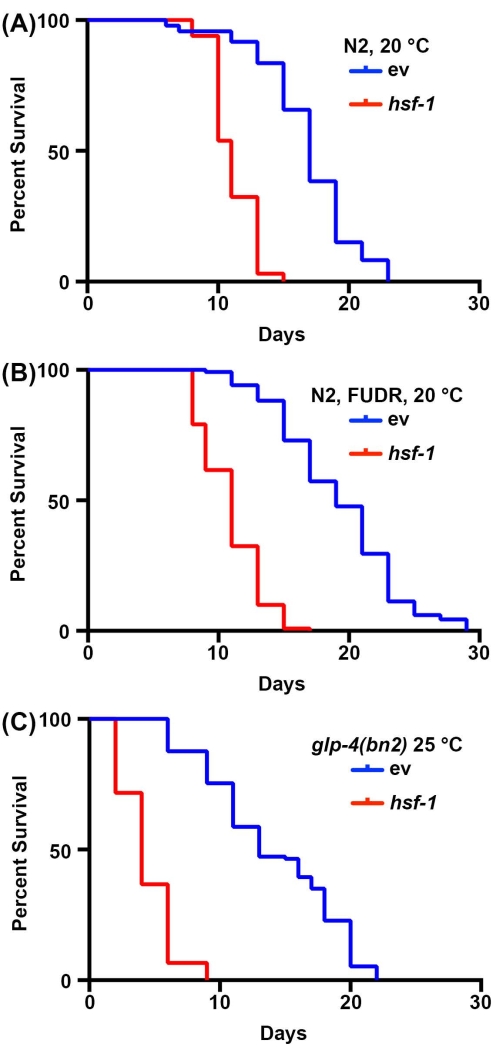
Figure 1: Comparison of lifespan measurements with and without sterilization. (A) Lifespans of wild-type N2 nematodes grown on NGM-agar plates seeded with empty vector (ev) or hsf-1 RNAi bacteria at 20 °C. Animals were moved away from progeny on days 1, 3, 5, and 7 of adulthood. (B) Lifespans of wild-type N2 nematodes grown on NGM-agar-FUDR plates seeded with empty vector (ev) or hsf-1 RNAi bacteria at 20 °C. Animals were grown to adulthood on standard ev or hsf-1 RNAi plates, and then moved to FUDR plates on day 1 of adulthood. (C) Lifespans of glp-4(bn2) mutant animals grown on NGM-agar plates seeded with empty vector (ev) or hsf-1 RNAi at 25 °C. For all conditions, animals were scored for death every 2 days until all animals were recorded as dead or censored. Animals with bagging, protrusion, or explosion of the vulva, or those that crawled up the sides of the plates and desiccated were censored. All statistics were performed using Log-Rank Mantel-Cox testing and can be found in Table 2. Please click here to view a larger version of this figure.
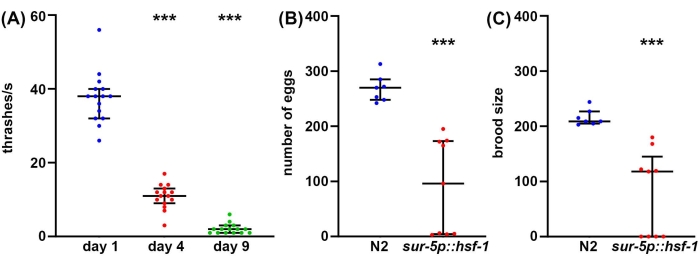
Figure 2: Egg count, brood size, and thrashing as measurements of healthspan. (A) Thrashing assays were performed on glp-4(bn2) mutant animals grown on NGM-agar plates seeded with empty vector at 25 °C on day 1 (blue), day 4 (red), and day 9 (green) animals. Thrashing was scored in animals placed into M9 solution on an NGM-agar plate, video recorded using a standard smartphone camera mounted onto an eyepiece of a standard dissecting scope, and thrashing scored in slow motion for accuracy. n = 15 animals per condition. (B) Egg counts were measured in wild-type N2 (blue) and sur-5p::hsf-1 (red) animals. Animals were grown at 20 °C and moved onto fresh plates, and eggs were counted every 12 h. The total number of eggs laid was summed. n = 7 animals for wildtype and 9 animals for sur-5p::hsf-1. (C) Brood assays were measured on the same animals as (B) where eggs were grown at 20 °C for 2 days to allow hatching, and all hatched eggs were counted. *** = p < 0.001 calculated using non-parametric Mann-Whitney testing. Each dot represents a single animal, and lines represent the median and interquartile range. Please click here to view a larger version of this figure.
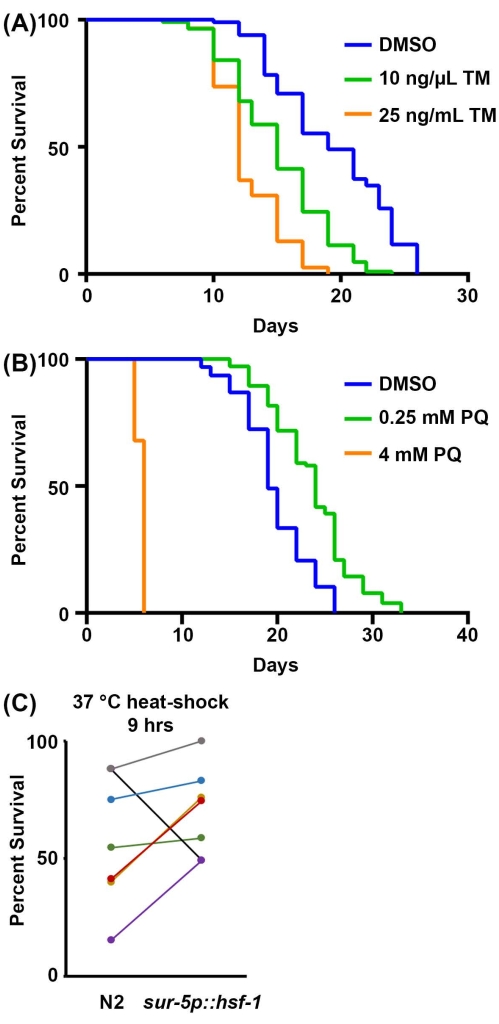
Figure 3: Stress resilience as a proxy to organismal health. (A) Survival assay of N2 animals grown on the empty vector RNAi bacteria at 20 °C. Animals were moved to plates containing either 1% DMSO, 10 ng/µL tunicamycin (TM), or 25 ng/µL TM on day 1 of adulthood. (B) Survival assay of N2 animals grown on the empty vector RNAi bacteria at 20 °C. Animals were grown from the hatch on plates containing either water, 0.25 mM paraquat (PQ), or 4 mM PQ. For A-B, animals were scored for death every 2 days until all animals were recorded as dead or censored. Animals with bagging, protrusion, or explosion of the vulva, or those that crawled up the sides of the plates and desiccated were censored. All statistics were performed using Log-Rank Mantel-Cox testing (Table 2). (C) Pooled data of all 37 °C thermotolerance assays for wild-type N2 animals versus overexpression of hsf-1 (sur-5p::hsf-1). Data are represented as percent alive at time = 9 h of a thermotolerance assay, with each line representing a matched experiment performed on the same day. Animals were grown on the empty vector RNAi bacteria at 20 °C and moved to 37 °C on day 1 of adulthood for assay. n = 60 animals per strain per replicate. Please click here to view a larger version of this figure.
| Reagent | Recipe | ||
| Bleach solution | 1.8% (v/v) sodium hypochlorite, 0.375 M KOH | ||
| Carbenicillin | 100 mg/mL stock solution (1000x) in water. Store at 4 °C for up to 6 months or -20 °C for long-term storage | ||
| FUDR | 10 mg/mL solution in water. Store at -20 °C. | ||
| IPTG | 1 M solution in water. | ||
| Lysogeny Broth (LB) | In this protocol, commercial LB was used (see Table of Materials), but all standard LB home-made recipes using Bacto-tryptone, yeast extract, and NaCl are sufficient. | ||
| M9 solution | 22 mM KH2PO4 monobasic, 42.3 mM Na2HPO4, 85.6 mM NaCl, 1 mM MgSO4 | ||
| Nematode Growth Media (NGM) | 1 mM CaCl2, 5 µg/mL cholesterol, 25 mM KPO4 pH 6.0, 1 mM MgSO4, 2% (w/v) agar, 0.25% (w/v) Bacto-Peptone, 51.3 mM NaCl | ||
| NGM RNAi plates | 1 mM CaCl2, 5 µg/mL cholesterol, 25 mM KPO4 pH 6.0, 1 mM MgSO4, 2% (w/v) agar, 0.25% (w/v) Bacto-Peptone, 51.3 mM NaCl, 1 mM IPTG, 100 µg/mL carbenicillin/ampicillin. Store at 4 ° C in dark for up to 3 months | ||
| NGM RNAi DMSO | 1 mM CaCl2, 5 µg/mL cholesterol, 25 mM KPO4 pH 6.0, 1 mM MgSO4, 2% (w/v) agar, 0.25% (w/v) Bacto-Peptone, 51.3 mM NaCl, 1 mM IPTG, 100 µg/mL carbenicillin/ampicillin; 1% DMSO | ||
| (control for tunicamycin) | |||
| NGM RNAi TM | 1 mM CaCl2, 5 µg/mL cholesterol, 25 mM KPO4 pH 6.0, 1 mM MgSO4, 2% (w/v) agar, 0.25% (w/v) Bacto-Peptone, 51.3 mM NaCl, 1 mM IPTG, 100 µg/mL carbenicillin/ampicillin; 1% DMSO, 25 ng/µL tunicamycin | ||
| Paraquat | 400 mM solution in water – should be prepared fresh | ||
| Tetracycline | 10 mg/mL stock solution (500x) in 100% ethanol. Store at -20 °C | ||
| Tunicamycin | 2.5 mg/mL stock solution in 100% DMSO. Store at -80 °C for long-term storage. This is a 100x solution (25 ng/µL working solution) | ||
Table 1. Recipes for reagents and media for protocols.
| Corresponding Figure panel | Strain, Treatment | Median Lifespan | # Deaths/# Total | p-value (Log-Rank) | |
| 1A | N2, vector RNAi, 20 °C | 17 | 74/120 | -- | |
| N2, hsf-1 RNAi, 20 °C | 11 | 65/120 | <0.001 | ||
| 1B | N2, vector RNAi, FUDR, 20 °C | 19 | 120/120 | -- | |
| N2, hsf-1 RNAi, FUDR, 20 °C | 11 | 116/120 | <0.001 | ||
| 1C | N2, glp-4(bn2), vector RNAi, 25 °C | 13 | 115/121 | -- | |
| N2, glp-4(bn2), hsf-1 RNAi, 25 °C | 4 | 120/120 | < 0.001 | ||
| 2A | N2, vector RNAi, 20 °C, 1% DMSO | 19 | 85/120 | -- | |
| N2, vector RNAi, 20 °C, 10 ng/µL tunicamycin | 15 | 109/120 | <0.001 | ||
| N2, vector RNAi, 20 °C, 25 ng/µL tunicamycin | 12 | 117/120 | <0.001 | ||
| 2B | N2, vector RNAi, 20 °C | 19 | 84/120 | -- | |
| N2, vector RNAi, 20 °C, 0.25 mM paraquat | 24 | 91/120 | <0.001 | ||
| N2, vector RNAi, 20 °C, 4 mM paraquat | 6 | 50/120 | <0.001 | ||
Table 2. Statistics for lifespan and stress resilience.
Discussion
Lifespan, most simply defined as the duration of life, is a clear binary phenomenon in most organisms-either an organism is living or is not. However, longevity does not always correlate with an organism's health. For example, mitochondrial hormesis models where exposure to mitochondrial stress dramatically increases lifespan are generally some of the longest-lived animals, yet exhibit stunted growth and decreased metabolic function37,54. Similarly, animals with hyperactive endoplasmic reticulum stress responses also exhibit certain behaviors and phenotypes that can be correlated with decreased health, despite having dramatically improved protein homeostasis and lifespan36,49. Finally, many longevity paradigms in model organisms including increased HSF-1 function55, increased XBP-1 function56, and altered FoxO signaling57 are all correlated with increased cancer risk, and it is inarguable that extended lifespan is not beneficial if an organism is in a constant struggle with cancer and other health maladies. Therefore, longevity cannot be a standalone measurement in aging biology.
Thus, the concept of healthspan has been a growing field in aging biology. Healthspan, loosely defined as the period of life that one is healthy, is more difficult to ascertain than longevity. However, unlike longevity, the concept of "health" is complicated, as there are many different readouts to organismal health: on the organismal level, there are muscle function/strength, neuronal/cognitive function, reproductive health, etc.; on the cellular level there are protein homeostasis, lipid homeostasis, glucose homeostasis, metabolism, etc. In 2014, aging biologists have definitively characterized biological hallmarks of aging with the structured definition that it must be something that naturally breaks down during aging and can experimentally be altered such that experimental exacerbation should accelerate aging and experimental intervention should slow down aging. These nine hallmarks of aging include genomic instability, telomere attrition, epigenetic alterations, loss of protein homeostasis (proteostasis), stem cell exhaustion, altered intercellular signaling, mitochondrial dysfunction, deregulated nutrient sensing, and cellular senescence58. Since then, numerous studies argue other factors should be included, including extracellular proteins and systemic physiology such as immunity and inflammation59. Ultimately, the complex definition of healthspan mandates that organismal health be measured using multiple different methods.
Therefore, in this manuscript, multiple methods are presented to measure various aspects of healthspan using the nematode model, C. elegans. We assay locomotory behavior using thrashing assays, reproductive health using egg count and brood size, and sensitivity to stress. Indeed, locomotory behavior is a gold-standard method for measuring healthspan, as organisms exhibit significant loss of motility and movement during aging51. Loss of locomotory behavior can be ascribed to multiple hallmarks of aging, as muscle function in C. elegans is dependent on proper proteostasis60, mitochondrial dysfunction61, and neuron-muscle signaling62. While this manuscript focuses on one measurement of locomotory behavior, it is important to note that many other methods exist, including motility of animals on a solid agar plate, response to touch51, and chemotaxis assays63. However, these methods generally require more sophisticated recording devices, usage of worm-tracking software, or usage of expensive, dangerous, or volatile chemicals, all of which may be prohibitive in some research settings.
In addition, assays for egg count and brood size are presented as a method of measuring reproductive health and as the simplest method to measure cell division in adult worms, since adult worms are post-mitotic and only germ cells and embryos undergo cell division within an adult worm64. As a measure of cell division, reproductive health can be relevant for the aging hallmarks of cellular senescence and stem cell exhaustion. Reproductive health can be affected by many factors, including pathogenic infection65 or exposure to stress49, though there is no direct correlation between reproductive health and longevity. In fact, some long-lived animals exhibit a significant decrease in brood size49, and it is even possible that there exists an inverse correlation between longevity and brood size50. This is not a phenomenon specific to C. elegans, as detrimental effects of reproduction on longevity have long been observed in humans66, companion dogs67, and mice68. Still, we provide egg count and brood size as a reliable and low-cost method for measuring reproductive health with the caveat that reproductive health may not correlate with longevity or healthspan.
Finally, survival assays are offered as an indirect measure of organismal health. Importantly, cellular stress responses, including response to thermal stress69 and ER stress35 rapidly decline during the aging process and have direct relevance to the aging hallmark of proteostasis70,71. In contrast, hyperactivating stress responses can significantly increase lifespan by promoting resilience to stress35,37,38. While this study focuses on the simplest and lowest cost methods, a large number of alternative methods for stress resilience assays exist for thermotolerance41 and oxidative stress66, each requiring a different set of equipment and consumables. Beyond simple exposure studies to stressors, other physiological methods can be performed depending on access to equipment. For example, an extracellular flux analyzer can monitor mitochondrial function and cellular respiration73; fluorescent dissection microscopes will allow measurements of transcriptional reporters for stress response activation20; and high-resolution compound or confocal microscopes can be used to measure organelle morphology with fluorescent probes for mitochondria74, the endoplasmic reticulum75,76, and actin cytoskeleton77.
As a final cautionary tale for measurements of longevity, while chemical and genetic methods for sterilizing worms are offered to significantly decrease cost, it is important to note that both can directly impact lifespan. For example, exposure to FUDR has been previously reported to impact both lifespan and thermotolerance45. And while the glp-4(bn2) mutant itself does not have any direct effects on lifespan, growth at 25 °C is a mild heat-stress33,34 and thus can impact lifespan2. There exists other methods for sterilizing C. elegans, including auxin-mediated sterility78 or alternative temperature-sensitive sperm-deficient mutants79. However, all methods have some caveats, and care should be taken to utilize the least detrimental assay for each laboratory's scientific needs. One final limitation of longevity studies is potential variability that can occur due to low sample sizes or simply by an objective error by the investigator. This can be circumvented as new technologies are born in automated lifespan technologies80, but again these systems are costly and require some engineering and computational equipment and skills. Ultimately, the collection of methods provided here is a reliable set of tools that can be quickly adapted and learned in almost any institution and provide a solid foundation for aging biology.
Disclosures
The authors declare no competing financial interests.
Acknowledgements
G.G. is supported by T32AG052374 and R.H.S. is supported by R00AG065200 from the National Institute on Aging. We thank the CGC (funded by NIH Office of Research Infrastructure Program P40 OD010440) for the strains.
Materials
| Name | Company | Catalog Number | Comments |
| APEX IPTG | Genesee | 18-242 | for RNAi |
| Bacto Agar | VWR | 90000-764 | for NGM plates |
| Bacto Peptone | VWR | 97064-330 | for NGM plates |
| Calcium chloride dihydrate | VWR | 97061-904 | for NGM plates |
| Carbenicillin | VWR | 76345-522 | for RNAi |
| Cholesterol | VWR | 80057-932 | for NGM plates |
| DMSO | VWR | BDH1115-1LP | solvent for drugs |
| LB Broth | VWR | 95020-778 | for LB |
| Magnesium sulfate heptahydrate | VWR | 97062-132 | for NGM plates, M9 |
| Paraquat | Sigma-Aldrich | 36541 | for oxidative/mitochondrial stress |
| Potassium Chloride | VWR | 97061-566 | for bleach soluton |
| Potassium phosphate dibasic | VWR | EM-PX1570-2 | for NGM plates |
| Potassium phosphate monobasic | VWR | EM-PX1565-5 | for M9 |
| S7E Dissecting Scope | Leica | 10450840 | Standard dissecting microscope |
| Sodium Chloride | VWR | EM-SX0420-5 | for NGM plates, M9 |
| Sodium hypochlorite | VWR | RC7495.7-32 | for bleach solution |
| Sodium phosphate dibasic | VWR | 71003-472 | for M9 |
| Tetracycline hydrochloride | VWR | 97061-638 | for RNAi |
| Tunicamycin | Sigma-Aldrich | T7765-50MG | for ER stress |
References
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics. 77 (1), 71-94 (1974).
- Klass, M. R. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mechanisms of Ageing and Development. 6 (6), 413-429 (1977).
- Friedman, D. B., Johnson, T. E. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 118 (1), 75-86 (1988).
- Kenyon, C., Chang, J., Gensch, E., Rudner, A., Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature. 366 (6454), 461-464 (1993).
- Lithgow, G. J., White, T. M., Melov, S., Johnson, T. E. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proceedings of the National Academy of Sciences of the United States of America. 92 (16), 7540-7544 (1995).
- Epel, E. S., Lithgow, G. J. Stress biology and aging mechanisms: toward understanding the deep connection between adaptation to stress and longevity. The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 69, 10-16 (2014).
- Luo, Y. Long-lived worms and aging. Redox Report: Communications in Free Radical Research. 9 (2), 65-69 (2004).
- Tissenbaum, H. A. Using C. elegans for aging research. Invertebrate Reproduction & Development. 59, 59-63 (2015).
- Zhang, S., Li, F., Zhou, T., Wang, G., Li, Z. Caenorhabditis elegans as a useful model for studying aging mutations. Frontiers in Endocrinology. 11, 554994 (2020).
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics. 77 (1), 71-94 (1974).
- Rual, J. -. F., et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Research. 14 (10), 2162-2168 (2004).
- Timmons, L., Court, D. L., Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263 (1-2), 103-112 (2001).
- Reboul, J., et al. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nature Genetics. 27 (3), 332-336 (2001).
- Lee, S. S., et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature Genetics. 33 (1), 40-48 (2003).
- Reinke, S. N., Hu, X., Sykes, B. D., Lemire, B. D. Caenorhabditis elegans diet significantly affects metabolic profile, mitochondrial DNA levels, lifespan and brood size. Molecular Genetics and Metabolism. 100 (3), 274-282 (2010).
- Revtovich, A. V., Lee, R., Kirienko, N. V. Interplay between mitochondria and diet mediates pathogen and stress resistance in Caenorhabditis elegans. PLOS Genetics. 15 (3), 1008011 (2019).
- Pang, S., Curran, S. P. Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metabolism. 19 (2), 221-231 (2014).
- Brooks, K. K., Liang, B., Watts, J. L. The influence of bacterial diet on fat storage in C. elegans. PLOS ONE. 4 (10), 7545 (2009).
- Soukas, A. A., Kane, E. A., Carr, C. E., Melo, J. A., Ruvkun, G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes & Development. 23 (4), 496-511 (2009).
- Bar-Ziv, R., et al. Measurements of physiological stress responses in C. Elegans. Journal of Visualized Experiments: JoVE. (159), e61001 (2020).
- Xiao, R., et al. RNAi interrogation of dietary modulation of development, metabolism, behavior, and aging in C. elegans. Cell Reports. 11 (7), 1123-1133 (2015).
- Calixto, A., Chelur, D., Topalidou, I., Chen, X., Chalfie, M. Enhanced neuronal RNAi in C. elegans using SID-1. Nature Methods. 7 (7), 554-559 (2010).
- Dickinson, D. J., Goldstein, B. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics. 202 (3), 885-901 (2016).
- Kim, H. -. M., Colaiácovo, M. P. CRISPR-Cas9-guided genome engineering in C. elegans. Current Protocols in Molecular Biology. 129 (1), 106 (2019).
- Farboud, B., Severson, A. F., Meyer, B. J. Strategies for efficient genome editing using CRISPR-Cas9. Genetics. 211 (2), 431-457 (2019).
- Transformation and Microinjection. WormBook: The Online Review of C. elegans Biology Available from: https://www.ncbi.nlm.nih.gov/books/NBK19648/ (2006)
- Frøkjaer-Jensen, C., et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature Genetics. 40 (11), 1375-1383 (2008).
- Kaymak, E., et al. Efficient generation of transgenic reporter strains and analysis of expression patterns in Caenorhabditis elegans using Library MosSCI. Developmental Dynamics: an Official Publication of the American Association of Anatomists. 245 (9), 925-936 (2016).
- Mariol, M. -. C., Walter, L., Bellemin, S., Gieseler, K. A rapid protocol for integrating extrachromosomal arrays with high transmission rate into the C. elegans genome. Journal of Visualized Experiments: JoVE. (82), (2013).
- Rastogi, S., et al. Caenorhabditis elegans glp-4 encodes a valyl aminoacyl tRNA synthetase. G3: Genes|Genomes|Genetics. 5 (12), 2719-2728 (2015).
- Beanan, M. J., Strome, S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 116 (3), 755-766 (1992).
- Santi, D. V., McHenry, C. S. 5-Fluoro-2′-Deoxyuridylate: covalent complex with thymidylate synthetase. Proceedings of the National Academy of Sciences. 69 (7), 1855-1857 (1972).
- Lithgow, G. J., White, T. M., Hinerfeld, D. A., Johnson, T. E. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. Journal of Gerontology. 49 (6), 270-276 (1994).
- Labbadia, J., Morimoto, R. I. The biology of proteostasis in aging and disease. Annual Review of Biochemistry. 84 (1), 435-464 (2015).
- Taylor, R. C., Dillin, A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 153 (7), 1435-1447 (2013).
- Higuchi-Sanabria, R., et al. Divergent nodes of non-autonomous UPRER signaling through serotonergic and dopaminergic neurons. Cell Reports. 33 (10), 108489 (2020).
- Durieux, J., Wolff, S., Dillin, A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 144 (1), 79-91 (2011).
- Morley, J. F., Morimoto, R. I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Molecular Biology of the Cell. 15 (2), 657-664 (2004).
- Heifetz, A., Keenan, R. W., Elbein, A. D. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry. 18 (11), 2186-2192 (1979).
- Castello, P. R., Drechsel, D. A., Patel, M. Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. The Journal of Biological Chemistry. 282 (19), 14186-14193 (2007).
- Park, H. -. E. H., Jung, Y., Lee, S. -. J. V. Survival assays using Caenorhabditis elegans. Molecules and Cells. 40 (2), 90-99 (2017).
- Gardner, M., Rosell, M., Myers, E. M. Measuring the effects of bacteria on C. Elegans behavior using an egg retention assay. Journal of Visualized Experiments: JoVE. (80), e51203 (2013).
- Waggoner, L. E., Hardaker, L. A., Golik, S., Schafer, W. R. Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics. 154 (3), 1181-1192 (2000).
- Zevian, S. C., Yanowitz, J. L. Methodological considerations for heat shock of the nematode Caenorhabditis elegans. Methods. 68 (3), 450-457 (2014).
- Feldman, N., Kosolapov, L., Ben-Zvi, A. Fluorodeoxyuridine improves Caenorhabditis elegans proteostasis independent of reproduction onset). PLOS ONE. 9 (1), 85964 (2014).
- Hsu, A. -. L., Murphy, C. T., Kenyon, C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 300 (5622), 1142-1145 (2003).
- Bansal, A., Zhu, L. J., Yen, K., Tissenbaum, H. A. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proceedings of the National Academy of Sciences of the United States of America. 112 (3), 277-286 (2015).
- Hodgkin, J., Barnes, T. M. More is not better: brood size and population growth in a self-fertilizing nematode. Proceedings. Biological Sciences. 246 (1315), 19-24 (1991).
- Ozbey, N. P., et al. Tyramine Acts Downstream of Neuronal XBP-1s to coordinate inter-tissue UPRER activation and behavior in C. elegans. Developmental Cell. 55 (6), 754-770 (2020).
- Lee, Y., et al. Inverse correlation between longevity and developmental rate among wild C. elegans strains. Aging. 8 (5), 986-994 (2016).
- Swierczek, N. A., Giles, A. C., Rankin, C. H., Kerr, R. A. High-throughput behavioral analysis in C. elegans. Nature Methods. 8 (7), 592-598 (2011).
- Lee, S. -. J., Hwang, A. B., Kenyon, C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Current Biology: CB. 20 (23), 2131-2136 (2010).
- Baird, N. A., et al. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. 346 (6207), 360-363 (2014).
- Houtkooper, R. H., et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497 (7450), 451-457 (2013).
- Carpenter, R. L., Gökmen-Polar, Y. HSF1 as a cancer biomarker and therapeutic target. Current Cancer Drug Targets. 19 (7), 515-524 (2019).
- Chen, S., et al. The emerging role of XBP1 in cancer. Biomedicine & Pharmacotherapy. 127, 110069 (2020).
- Yadav, R. K., Chauhan, A. S., Zhuang, L., Gan, B. FoxO transcription factors in cancer metabolism. Seminars in Cancer Biology. 50, 65-76 (2018).
- López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., Kroemer, G. The hallmarks of aging. Cell. 153 (6), 1194-1217 (2013).
- Kennedy, B. K., et al. Geroscience: linking aging to chronic disease. Cell. 159 (4), 709-713 (2014).
- Ben-Zvi, A., Miller, E. A., Morimoto, R. I. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proceedings of the National Academy of Sciences. 106 (35), 14914-14919 (2009).
- Hewitt, J. E., et al. Muscle strength deficiency and mitochondrial dysfunction in a muscular dystrophy model of Caenorhabditis elegans and its functional response to drugs. Disease Models & Mechanisms. 11 (12), (2018).
- Gao, S., Zhen, M. Action potentials drive body wall muscle contractions in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 108 (6), 2557-2562 (2011).
- Margie, O., Palmer, C., Chin-Sang, I. C. elegans chemotaxis assay. Journal of Visualized Experiments: JoVE. (74), e50069 (2013).
- Flemming, A. J., Shen, Z. Z., Cunha, A., Emmons, S. W., Leroi, A. M. Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proceedings of the National Academy of Sciences of the United States of America. 97 (10), 5285-5290 (2000).
- Madhu, B. J., Salazar, A. E., Gumienny, T. L. Caenorhabditis elegans egg-laying and brood-size changes upon exposure to Serratia marcescens and Staphylococcus epidermidis are independent of DBL-1 signaling. microPublication Biology. 2019, (2019).
- Westendorp, R. G., Kirkwood, T. B. Human longevity at the cost of reproductive success. Nature. 396 (6713), 743-746 (1998).
- Hoffman, J. M., Creevy, K. E., Promislow, D. E. L. Reproductive capability is associated with lifespan and cause of death in companion dogs. PLOS ONE. 8 (4), 61082 (2013).
- Garratt, M., Try, H., Smiley, K. O., Grattan, D. R., Brooks, R. C. Mating in the absence of fertilization promotes a growth-reproduction versus lifespan trade-off in female mice. Proceedings of the National Academy of Sciences of the United States of America. 117 (27), 15748-15754 (2020).
- Labbadia, J., Morimoto, R. I. Repression of the heat shock response is a programmed event at the onset of reproduction. Molecular Cell. 59 (4), 639-650 (2015).
- Higuchi-Sanabria, R., Frankino, P. A., Paul, J. W., Tronnes, S. U., Dillin, A. A futile battle? Protein quality control and the stress of aging. Developmental Cell. 44 (2), 139-163 (2018).
- Dutta, N., Garcia, G., Higuchi-Sanabria, R. Hijacking cellular stress responses to promote lifespan. Frontiers in Aging. 3, (2022).
- Senchuk, M. M., Dues, D. J., Van Raamsdonk, J. M. Measuring oxidative stress in Caenorhabditis elegans: paraquat and juglone sensitivity assays. Bio-protocol. 7 (1), 2086 (2017).
- Leung, D. T. H., Chu, S. Measurement of oxidative stress: mitochondrial function using the seahorse system. Methods in molecular biology. 1710, 285-293 (2018).
- Daniele, J. R., et al. High-throughput characterization of region-specific mitochondrial function and morphology. Scientific Reports. 7 (1), 6749 (2017).
- Xu, N., et al. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. The Journal of Cell Biology. 198 (5), 895-911 (2012).
- Daniele, J. R., et al. UPRER promotes lipophagy independent of chaperones to extend life span. Science Advances. 6 (1), 1441 (2020).
- Higuchi-Sanabria, R., et al. Spatial regulation of the actin cytoskeleton by HSF-1 during aging. Molecular Biology of the Cell. 29 (21), 2522-2527 (2018).
- Kasimatis, K. R., Moerdyk-Schauwecker, M. J., Phillips, P. C. Auxin-mediated sterility induction system for longevity and mating studies in Caenorhabditis elegans. G3: Genes|Genomes|Genetics. 8 (8), 2655-2662 (2018).
- Fabian, T. J., Johnson, T. E. Production of age-synchronous mass cultures of Caenorhabditis elegans. Journal of Gerontology. 49 (4), 145-156 (1994).
- Stroustrup, N., et al. The Caenorhabditis elegans lifespan machine. Nature Methods. 10 (7), 665-670 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved