Method Article
Fixation of Embryonic Mouse Tissue for Cytoneme Analysis
In This Article
Summary
Here, an optimized step-by-step protocol is provided for fixation, immunostaining, and sectioning of embryos to detect specialized signaling filopodia called cytonemes in developing mouse tissues.
Abstract
Developmental tissue patterning and postdevelopmental tissue homeostasis depend upon controlled delivery of cellular signals called morphogens. Morphogens act in a concentration- and time-dependent manner to specify distinct transcriptional programs that instruct and reinforce cell fate. One mechanism by which appropriate morphogen signaling thresholds are ensured is through delivery of the signaling proteins by specialized filopodia called cytonemes. Cytonemes are very thin (≤200 nm in diameter) and can grow to lengths of several hundred microns, which makes their preservation for fixed-image analysis challenging. This paper describes a refined method for delicate handling of mouse embryos for fixation, immunostaining, and thick sectioning to allow for visualization of cytonemes using standard confocal microscopy. This protocol has been successfully used to visualize cytonemes that connect distinct cellular signaling compartments during mouse neural tube development. The technique can also be adapted to detect cytonemes across tissue types to facilitate the interrogation of developmental signaling at unprecedented resolution.
Introduction
Embryonic development is orchestrated through coordinated activation of morphogen signaling pathways. Morphogens are small, secreted proteins that are categorized into Sonic Hedgehog (SHH), transforming growth factor β (TGF-β)/bone morphogenic protein (BMP), Wingless-related integration site (WNT), and fibroblast and epidermal growth factor (FGF/EGF) families. Morphogens are produced and released from cellular organizing centers during tissue development and establish signaling gradients across organizing fields of cells to inform tissue morphogenesis1,2,3,4,5. One representation of morphogen gradients is found in the developing nervous system, where the presumptive central nervous system is patterned through morphogen pathway activation. This tissue, referred to as the neural tube, is comprised of opposing gradients of SHH secreted by the ventral-most notochord and floor plate, and WNTs/BMPs secreted from the dorsal roofplate, to pattern distinct neural progenitor regions6. The neural tube is commonly used to interrogate morphogen gradient integrity in developmental research.
Morphogen gradient formation relies on tight regulation of signal dispersion7. One cellular mechanism by which this occurs is through the formation of long signaling filopodia called cytonemes that facilitate direct delivery of morphogens from signal-producing cells to specific target cell populations. Cytonemes have been observed to extend hundreds of micrometers to deposit morphogens on signal-receiving cell membranes8,9. Disruption of cytoneme-mediated morphogen transport leads to developmental anomalies in both flies and vertebrates, highlighting their importance during tissue patterning10,11,12,13,14.
To date, cytonemes have been documented in Drosophila, chick, and zebrafish models, but imaging of the structures in developing mammalian embryos remains challenging8,9,15. A hurdle to effectively imaging cytonemes in complex mammalian tissues in situ is their thin and fragile nature, which makes them susceptible to damage by conventional fixation methods8. We had previously developed and optimized protocols for a modified electron microscopy fixative (MEM-fix) to preserve cytonemes in cultured cells and enable their study using confocal microscopy16,17.
Use of the MEM-fix technique has permitted identification of some of the molecules involved in SHH-induced cytoneme formation and function11,16,17. However, confirmation of these findings in the physiologically relevant context of neural tube patterning necessitated the development of novel techniques to fix and image mouse embryonic tissue. A protocol to fix mouse embryos in a manner that maintains cytoneme integrity and permits immunostaining and subsequent sectioning of embryonic tissue for confocal analysis is outlined here. This protocol was developed using a membrane-tethered green fluorescent protein (GFP) to label membrane extensions from the SHH-producing cells in the developing neural tube. Implementation of this protocol will address unanswered questions pertaining to the prevalence and significance of cytonemes in developing mammalian systems.
Protocol
This protocol follows the approved animal care guidelines of Institutional Animal Care and Use Committee (IACUC) and St. Jude Children's Research Hospital. All strains were backcrossed five generations to the C57BL/6J strain.
1. Embryo isolation and whole mount staining
- Breed 6-week-old females and monitor for the presence of vaginal plug.
- Euthanize the pregnant dam by CO2 inhalation in a CO2 chamber followed by cervical dislocation according to the AVMA guidelines18. Make a Y-incision to the peritoneal cavity using dissecting scissors and forceps. Excise the uterus containing the E9.5 embryos as per approved institutional guidelines.
- Dissect the embryos in complete growth medium (Dulbecco's Modified Eagle Medium, supplemented with nonessential amino-acids, Na-pyruvate, L-glutamine, and 10% fetal bovine serum). Use forceps to remove the yolk sac, placenta, and surrounding membranes.
NOTE: When required, save each yolk sac for embryo genotyping. - Rinse the isolated embryos in Hank's balanced salt solution (HBSS) to remove any residual amniotic tissue and blood.
- Prepare the fixative. Add paraformaldehyde (PFA) to HBSS for a final working concentration of 4% PFA.
CAUTION: PFA is a toxic chemical, so inhalation or direct skin exposure must be avoided. Preparation of the fixative solution is performed under a fume hood with the proper personal protective equipment of gloves and a lab coat. - Add 1 mL of fixative to each well of a 24-well plate and place each embryo in an individual well. Incubate the embryos in fixative for 45 min with gentle agitation on a rocker.
NOTE: Critical: All washes and incubations must be done with gentle agitation (maximum 20 RPM) on a rocker or circular shaker, as abrupt handling or movement of the embryos will destroy the fixed cytonemes. Use a pipette to gently remove all solutions. - Remove the fixative and wash the embryos 3 x 30 min in Phosphate-Buffered Saline (PBS) with Ca2+ and Mg2+ with added 0.1% Triton.
- After washing, incubate the embryos in blocking solution (PBS with Ca2+ and Mg2+, 0.1% Triton, and 5% goat serum) with gentle agitation. Block 2 x 1 h. After the second blocking incubation, perform one quick rinse of the embryos using fresh blocking solution.
- During the second blocking step, prepare the primary antibody solution11. Dilute the antibodies to the optimized concentration in PBS with Ca2+ and Mg2+, 0.1% Tween-20, and 5% goat serum.
NOTE: For enhanced visualization of membrane GFP, chicken anti-GFP (1:250) can be used. - Remove the blocking solution and add 1 mL of primary antibody solution to each well. Incubate at 4 °C with gentle rotation for 3 days.
- Following the primary antibody incubation, wash the embryos 5 x 1 h at 20 RPM on a rocker in PBS with Ca2+ and Mg2+, 0.1% Tween-20, and 5% goat serum.
- Prepare the secondary antibody solution using F(ab')2 fragment secondaries at a 1:1,000 dilution in PBS with Ca2+ and Mg2+, 0.1% Tween-20, and 5% goat serum.
NOTE: Use of F(ab')2 fragments greatly enhances antibody penetration into sample. - Add 1 mL of secondary antibody solution to each well. Incubate with gentle rocking at 4 °C in the dark for 3 days.
NOTE: Important: From this point on, minimize embryo exposure to direct light. Optional: To prevent bacterial growth, add 0.2% sodium azide to the secondary antibody solution. - Remove the secondary antibody solution and wash the embryos 3 x 30 min in PBS with Ca2+ and Mg2+, 0.1% Tween-20, and 5% goat serum. If not performing actin staining with phalloidin or other actin dyes, after the first wash, add 4′,6-diamidino-2-phenylindole (DAPI) to PBS with Ca2+ and Mg2+, 0.1% Tween-20, and 5% goat serum and incubate for 1 h, followed by 3 x 30 min washes as described above.
NOTE: At this point, the embryos can be stored overnight at 4 °C in PBS or HBSS in the dark until the embedding step the following day. However, it is recommended that embryos are embedded and sectioned immediately.
2. Embryo embedding, sectioning, and mounting
- Prepare a 4% w/v Low Melting Point (LMP) agarose solution dissolved in HBSS or PBS with Ca2+ and Mg2+ to a final volume of ~3 mL per embryo. Add the appropriate weight of LMP agarose to HBSS or PBS with Ca2+ and Mg2+ and microwave until the LMP agarose is dissolved.
NOTE: Once the LMP agarose is dissolved, store the solution in a bead bath, incubator, or water bath set to 55 °C to prevent the LMP agarose solution from solidifying. - Use a 12-well plate as the embedding mold for embryos. Place the 12-well plate in the 55 °C bead bath. Add 2.5-3 mL of 4% LMP agarose to each well that will hold an embryo.
NOTE: Although other molds may be used, 12-well plate volumes are optimal for preparing the agarose block at later stages for sectioning. - Using a perforated spoon, transfer the embryos into individual wells containing 4% LMP agarose solution (Figure 1A).
- Transfer the 12-well plate from the bead bath to a benchtop. Use pipette tips to gently embed and orient the embryo so it is centered within the solution. Once the embryos are oriented, place the plate at -20 °C for 10 min to allow quick solidification of the block.
CRITICAL: Ensure the embryo is positioned in the center of the block. Embryos that sink to the bottom or are too close to the edge of the mold will likely be dislodged from the agarose block while sectioning.
- Transfer the 12-well plate from the bead bath to a benchtop. Use pipette tips to gently embed and orient the embryo so it is centered within the solution. Once the embryos are oriented, place the plate at -20 °C for 10 min to allow quick solidification of the block.
- Remove the entire agarose block from the well using a scalpel and cut a rectangular block around the embryo leaving ~0.3 cm of block on each side. Allow extra length along the caudal end of the embryo. When mounted on the vibratome, orient the embryo in an upright position in the upper section of the block (Figure 1B).
- Apply a strip of tape to the specimen holder of the vibratome and superglue the agarose block to the tape, oriented so the blade will generate axial sections of the embryo in an anterior (cranial) to posterior sequence.
- Fill the vibratome chamber with cold HBSS to ensure the sample is fully immersed, and then surround the chamber with ice.
- Set the vibratome speed to 0.2 mm/s and frequency between 5 and 7 (50-70 Hz) with cut thickness set to 100 µm. Perform serial axial sectioning of the embryo.
NOTE: Use a slow speed for sectioning. If the cut speed is increased above 0.25 mm/s, sectioning may tear the embryo or dislodge the embryo from the block.- During the sectioning series, use forceps to gently transfer individual sections to a separate 60 mm dish filled with HBSS. Use forceps to grab the block, not the tissue, to avoid tissue damage and destruction of cytonemes.
NOTE: Tissue sections should remain within the agarose block. If the tissue falls out of the block into the vibratome chamber, gently transfer by lifting the section out. Do not grab or pinch the tissue. CRITICAL: Any folding of tissue sections or abrupt handling will destroy fixed cytonemes in the tissue sections.
- During the sectioning series, use forceps to gently transfer individual sections to a separate 60 mm dish filled with HBSS. Use forceps to grab the block, not the tissue, to avoid tissue damage and destruction of cytonemes.
- If not performing F-actin staining, proceed to step 2.10.
- To perform F-actin staining, remove HBSS and incubate sections for 40 min with Actin-Red and DAPI solution diluted in PBS with Ca2+ and Mg2+, 0.1% Tween-20, and 5% goat serum at room temperature.
- Wash sections 3 x 20 min in PBS with Ca2+ and Mg2+ and 0.1% Tween-20.
- Using a hydrophobic marker pen, draw a hydrophobic barrier around the edges of a charged microscope slide and add a small volume of HBSS to fill the area.
- Use forceps or a perforated spoon to transfer sections to the slide.
NOTE: Any tissue sections that are not encased in agarose can be transferred via a transfer pipette. - Remove excess agarose block using forceps.
- Once all sections have been transferred to the slide, remove excess liquid via pipetting and the corner of an absorbent towelette, and then add several drops of mounting medium to the slide. Use enough mounting medium to cover the entire coverslip area when cured. Mount the coverslip by gently placing it on the slide.
NOTE: Avoid applying pressure or disturbing coverslip until the mounting medium has cured.
3. Imaging
- Perform imaging of tissue sections on any confocal or higher resolution microscope11. Analyze a minimum of three embryos per genotype.
NOTE: Images of tissue sections were acquired using a TCS SP8 STED 3x confocal microscope, followed by LIGHTNING deconvolution.
Results
Use of a larger mold of a 12-well plate with 2.5-3 mL of agarose solution per well was ideal for embedding and suspending several embryos over a short period of time (Figure 1A). The excess area allows for correct orientation when cutting the agarose block for sectioning. When cutting the agarose block, it is important to maintain excess agarose along the bottom of the block where it will be glued to the tape on the object holder. The embryo should be in the upper half of the block (Figure 1B). However, the bottom section should not be too large as excess block increases the chances of altering the cut angle as the blade pushes into the block while sectioning. Examples of correctly oriented sections are shown (Figure 1C,D).
While developing this protocol, vibratome sectioning was compared to cryostat sectioning. Cryostat sectioning of the tissue rarely preserved cellular extensions (Figure 2 cryostat and Figure 3A,B vibratome). Cryostat sections allowed for the detection of some GFP-positive membrane fragments between cells of the notochord and neural tube (Figure 2A arrowhead) and between adjacent neural tube cells (Figure 2B,B' arrowheads). However, F-actin staining of cellular extensions in the highly filamentous mesenchymal cells surrounding the neural tube was impaired in cryostat sections (Figure 2C,C' arrow, vs Figure 3C,D). These cryostat results indicate that only some traces of broken cellular extensions remain following this method. Thus, vibratome sectioning is preferred to efficiently preserve these delicate extensions for subsequent analysis.
Minimal disruption of whole embryo and individual tissue sections is essential for high-quality imaging of cellular extensions. Tissue sections that have undergone any folding or buckling will be evident by the absence of the notochord or a large separation (>30 µm) between the notochord and the ventral floor plate of the neural tube (Figure 3B). This is usually accompanied by loss of mesenchymal cells that normally surround the neural tube (Figure 3A, arrow). Damaged sections will also result in a loss of visible cellular membrane extensions migrating between epithelial cells (Figure 3B compared to Figure 3A, arrowheads). Even minor distortions to the sections may cause fragmentation of actin-based extensions and large gaps to form between cells (Figure 3D, arrowheads, compared to well-preserved Figure 3C), underscoring the need for delicate handling at all steps.
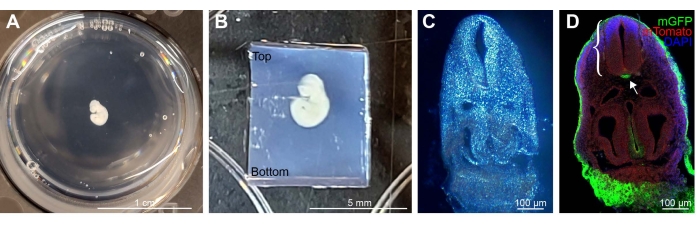
Figure 1: Example of E9.5 Shh-Cre; Rosa26 mTmG stained, embedded, and sectioned embryo. (A) A single embryo in a 12-well plate, embedded in 4% LMP agarose. (B) Example of a correctly oriented embryo within the agarose block cut down to size for vibratome mounting. Excess agarose block is present along the bottom. (C) Bright field image of 100 µm thick embryo section embedded in LMP agarose. (D) Immunofluorescence imaging of a section after removal of agarose. Anti-GFP-stained mGFP (green) represents Shh-expressing cells in the tissue, with other cell lineages expressing membrane Tomato (red), DAPI in blue. Neural tube (bracket) and mGFP-positive notochord (arrow) are clearly visible. Scale bars = 1 cm (A), 5 mm (B), and 100 µm (C,D). Abbreviations: LMP = low melting point; mGFP = membrane green fluorescent protein; Shh = Sonic Hedgehog; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
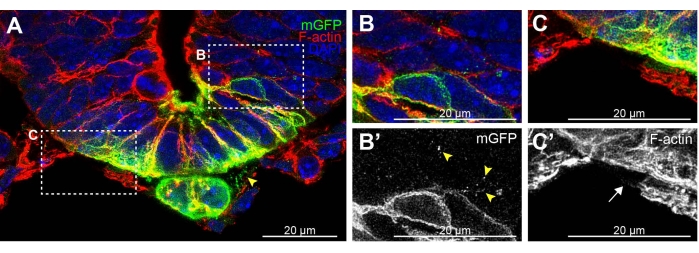
Figure 2: Example of cryostat-sectioned neural tube floor plate and notochord with fragmented membrane extensions. (A) 20 µm thick E9.5 Shh-Cre; Rosa26 mTmG19,20,21 section stained for GFP (green), F-actin (red), and DAPI (blue). mGFP puncta are visible between the notochord and floorplate (arrowhead) and (B,B') migrating between adjacent cells of the neural tube. (C,C') F-actin staining fails to detect any clear cytonemes on mesenchymal cells surrounding the neural tube (arrow). Scale bars = 20 µm. Abbreviations: mGFP = membrane green fluorescent protein; Shh = Sonic Hedgehog; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
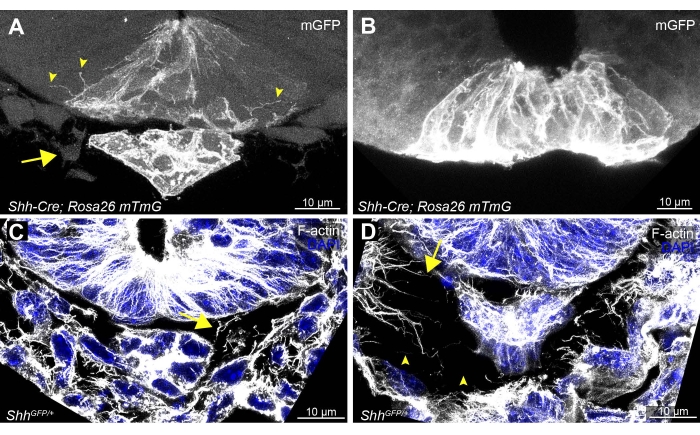
Figure 3: Examples of optimal and suboptimal vibratome tissue sections and staining of the neural tube floor plate, notochord, and surrounding cells. Examples of delicately handled sections (A,C), compared to (B) a folded tissue section and (D) a poorly handled section from a Shh-Cre; Rosa26 mTmG and a ShhGFP/+ embryo19,20,21. Optimally handled sections allow detection of cytonemes between adjacently localized floor plate neural epithelial cells (A, arrowheads). Neural tube adjacent notochord (A, mGFP-expressing) and mesenchymal cells (A, arrow) should be visible. (C,D) F-actin- and DAPI-stained sections should have consistent spacing of mesenchymal cells and F-actin-based cytonemes (arrows) surrounding the neural tube and notochord (C). Any minor folding or disruption of the sections can cause gaps and broken F-actin fragments (D, arrowheads). Scale bars = 10 µm. Abbreviations: mGFP = membrane green fluorescent protein; Shh = Sonic Hedgehog; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
Discussion
This protocol was developed for the preservation of delicate cytonemes in embryonic tissue sections for high-resolution fluorescence microscopy. To date, visualization of cytonemes in tissue across various model organisms has relied largely upon ectopic expression of fluorescently labeled proteins using live-tissue imaging. Unfortunately, the use of fluorescently labeled live-imaging is rarely conducive to the analysis of endogenously expressed proteins, can introduce time constraints, and requires specialized imaging stages and microscopes. The staining and sectioning method described here preserves cytonemes and other cellular extensions to permit immunohistochemistry for the eventual detection of endogenously expressed proteins. This technology will facilitate new insights into how morphogens are transported across different tissues in vivo and expands the scope of model systems where cytonemes can be studied.
This protocol uses whole-mount staining and vibratome thick-sectioning, which avoids the use of equilibration solution such as glycerol or cryoprotectant solutions such as sucrose. It aims to minimize sectioned tissue handling to allow for the maximal preservation of cellular extensions. Reduced handling of the tissue after sectioning provided consistently better results in the overall preservation of cytonemes. Thus, sectioning of the embryo is one of the final steps prior to imaging.
MEM-fix, a fixation technique developed for the preservation of cytonemes of cultured cells, consists of a combination of glutaraldehyde and PFA that allows for improved 3D preservation of the innate cell architecture comparable to their live dimensions16,17. Unfortunately, MEM-fix was not useful for embryo fixation because glutaraldehyde can auto-fluoresce and severely limits antibody penetration and epitope binding in cells due to high protein cross-linking activity22,23. Thus, sectioning protocols post PFA fixation conditions were optimized to allow for the preservation of cellular extensions in embryonic tissue. Although PFA can preserve cytoneme-like extensions in embryonic tissue, image analysis should be performed with caution. PFA can cause partial dehydration of individual cells and tissues, leading to minor volume reduction and the formation of small extracellular spaces within the fixed tissue.
This protocol avoids the extra steps of sucrose resaturation for cryoprotection and tissue clearing because sucrose or glycerol solutions can cause a minor resaturation of the tissue24. The resulting minor swelling can destroy fixed cellular extensions and cytonemes migrating between cells and across tissues. This was evident when comparing thick-sectioned vibratome to cryostat sections. Although increasing tissue thickness improved the preservation of intact cytonemes, the sucrose-resaturated cryostat sections consistently had a lower incidence of detectable cytonemes.
Optimization of this protocol revealed that 100 µm thick sections were the best for ensuring preservation of intact cytonemes. Thinner sections are typically used for imaging because most confocal microscopes are only capable of imaging at sufficient resolution to visualize cellular extensions to a depth of ~20-40 µm without clearing25. However, the mechanical forces and distortion applied to embryonic cells from the blade while cutting thin sections destroys cytonemes. Thicker sections allow for a larger area of depth within the section while preserving intact extensions within the tissue. Additionally, the use of thicker sections allows for ease of handling to prevent excessive folding or buckling of the tissue sections.
This protocol was optimized for the maximum preservation of cytonemes in tissue of E8-10.5 mouse embryos. Minimal manipulation of the tissue after sectioning provided consistently improved overall preservation of cytonemes. It is likely that further optimization will be required to adapt the technique for later developmental stages and adult tissue sections due to increased size and tissue complexity. This will require sectioning followed by immunofluorescence staining. Such tissue may require additional clearing steps and adaptation of tissue handling during sectioning to preserve cytoneme-like extensions. These additional steps will need to be considered and addressed for future applications of this protocol.
Disclosures
The authors have no competing interests to declare.
Acknowledgements
Images were acquired using microscopes maintained by the Cell and Molecular Biology Imaging Core at St. Jude Children's Research Hospital. Mouse strains were obtained from JAX. This work was supported by National Institutes of Health grant R35GM122546 (SKO) and by ALSAC of St. Jude Children's Research Hospital.
Materials
| Name | Company | Catalog Number | Comments |
| 12-well plate; Nunc Cell-Culture Treated | Thermo Scientific | 150628 (Fisher Scientific 12-565-321) | |
| 24-Well Plate, Round Nunc | Thermo Fisher Scientific | 142475 | |
| 60 mm dish | Corning, Falcon | 353004 (Fisher Scientific 08772F) | |
| Absorbant towels (Professional Kimtech Science Kimwipes) | Kimberly-Clark KIMTECH | 34155 (Fisher Scientific 06666A) | |
| Actin Red 555 ReadyProbes Reagent | Invitrogen | R37112 | |
| Alexa Fluor 488 AffiniPure F(ab')2 Fragment Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson Immunoresearch Lab Inc | 703-546-155 | 1:1,000 working concentration |
| Bead Bath 6L 230V | Lab Armor | Item #12L048 (Mfr. Model #74220-706) | set to 55 °C |
| chicken anti-GFP | Aves Labs | SKU GFP-1020 | 1:250 working concentration |
| CO2 chamber | plexiglass chamber connected to a CO2 emitting line. | ||
| Confocal laser-scanning microscope | Leica Microsystems | model: Leica TCS SP8 | |
| DAPI Solution (1 mg/mL) | Thermo Fisher Scientific | 62248 | |
| Dissecting scissors (Vannas Scissors) | World Precision Instruments | 14003 | |
| Dulbecco's Modified Eagle Medium, high glucose, no glutamine | Thermo Fisher Scientific, Gibco | 11960044 | |
| Eppendorf Research Plus single channel pipette, 0.1-2.5 µL | Eppendorf | 3123000012 | |
| Eppendorf Research Plus single channel pipette, 0.5-10 µL | Eppendorf | 3123000020 | |
| Eppendorf Research Plus single channel pipette, 100-1,000 µL | Eppendorf | 3123000063 | |
| Eppendorf Research Plus single channel pipette, 20-200 µL | Eppendorf | 3123000055 | |
| Feather Disposable Scalpel #10 | Fisher Scientific | Catalog No.NC9999403 | |
| Fetal Bovine Serum, certified, United States | Thermo Fisher Scientific, Gibco | 16000044 | |
| Fisherbrand Fine Point High Precision Forceps | Fisher Scientific | 22-327379 | |
| Fisherbrand Labeling Tape | Fisher Scientific | 15-954 | |
| Formaldehyde, 16%, methanol free, Ultra Pure EM Grade | Polysciences | 18814-10 | |
| Hank's Balanced Salt Solution (HBSS) | Thermo Fisher Scientific, Gibco | 14025 | |
| ImmEdge Hydrophobic Barrier Pap Pen | Vector laboratories | H-4000 | |
| Leica Application Suite X (LAS X) with LIGHTNING | Leica Microsystems | Microscope software | |
| L-Glutamine (200 mM) | Thermo Fisher Scientific, Gibco | 25030081 | |
| Low Melting Point Agarose- UltraPure | Invitrogen | 16520 | |
| MEM Non-Essential Amino Acids Solution (100x) | Thermo Fisher Scientific, Gibco | 11140050 | |
| Moria MC 17 BIS Perforated Spoon | Fine Science Tools | 1037018 | |
| Mounting media (ProLong Diamond Antifade Mountant) | Thermo Fisher Scientific, Invitrogen | P36961 | |
| Mouse: B6.129X1(Cg)-Shhtm6Amc/J (ShhGFP/+) | The Jackson Laboratory | Strain #:008466 | |
| Mouse: B6.Cg-Shhtm1(EGFP/cre)Cjt/J (Shh-Cre) | The Jackson Laboratory | Strain #:005622 | |
| Mouse: C57BL/6J (B6) | The Jackson Laboratory | Strain #:000664 | |
| Mouse: Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (Rosa26 mTmG) | The Jackson Laboratory | Strain #:007576 | |
| Normal Goat Serum | Jackson ImmunoResearch | 005-000-121 (Fisher Scientific NC9660079) | |
| PBS + Ca2+ + Mg2+ | Thermo Fisher Scientific, Gibco | 14040133 | |
| Premium Microcentrifuge Tubes: 1.5 mL | Fisher Scientific | 05-408-129 | |
| SHARP Precision Barrier Tips, Extra Long for P-10 and Eppendorf 10 µL | Denville Scientific | P1096-FR | |
| SHARP Precision Barrier Tips, For P-1000 and Eppendorf 1,000, 1,250 µL | Denville Scientific | P1126 | |
| SHARP Precision Barrier Tips, For P-20 and Eppendorf 20 µL | Denville Scientific | P1121 | |
| SHARP Precision Barrier Tips, For P-200 and Eppendorf 200 µL | Denville Scientific | P1122 | |
| Sodium Azide | Sigma-Aldrich | S8032 | |
| Sodium Pyruvate (100 mM) (100x) | Thermo Fisher Scientific, Gibco | 11360070 | |
| Super Glue | Gorilla | ||
| SuperFrost Plus microscope slides | Fisher Scientific | 12-550-15 | |
| TPP centrifuge tubes, volume 15 mL, polypropylene | TPP Techno Plastic Products | 91015 | |
| TPP centrifuge tubes, volume 50 mL, polypropylene | TPP Techno Plastic Products | 91050 | |
| Transfer pipette, polyethylene | Millipore Sigma | Z135003 | |
| Triton X-100 | Sigma-Aldrich | T9284 | |
| Tween-20 | Sigma-Aldrich | P1379 | |
| Vari-Mix Platform Rocker | Thermo Fisher Scientific | 09-047-113Q | set to 20 RPM |
| Vibratome | Leica Biosytems | VT1000 S |
References
- Crick, F. Diffusion in embryogenesis. Nature. 225 (5231), 420-422 (1970).
- Zeng, X., et al. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature. 411 (6838), 716-720 (2001).
- Yu, S. R., et al. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 461 (7263), 533-536 (2009).
- Zhou, S., et al. Free extracellular diffusion creates the Dpp morphogen gradient of the Drosophila wing disc. Current Biology. 22 (8), 668-675 (2012).
- Ribes, V., Briscoe, J. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harbor Perspectives in Biology. 1 (2), 002014(2009).
- Le Dréau, G., Martí, E. Dorsal-ventral patterning of the neural tube: a tale of three signals. Developmental Neurobiology. 72 (12), 1471-1481 (2012).
- Kornberg, T. B., Guha, A. Understanding morphogen gradients: a problem of dispersion and containment. Current Opinion in Genetics & Development. 17 (4), 264-271 (2007).
- Ramirez-Weber, F. A., Kornberg, T. B. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 97 (5), 599-607 (1999).
- Sanders, T. A., Llagostera, E., Barna, M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 497 (7451), 628-632 (2013).
- Bischoff, M., et al. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nature Cell Biology. 15 (11), 1269-1281 (2013).
- Hall, E. T., et al. Cytoneme delivery of sonic hedgehog from ligand-producing cells requires Myosin 10 and a dispatched-BOC/CDON co-receptor complex. eLife. 10, 61432(2021).
- Huang, H., Kornberg, T. B. Cells must express components of the planar cell polarity system and extracellular matrix to support cytonemes. eLife. 5, 18979(2016).
- Brunt, L., et al. Vangl2 promotes the formation of long cytonemes to enable distant Wnt/β-catenin signaling. Nature Communications. 12 (1), 2058(2021).
- Roy, S., Huang, H., Liu, S., Kornberg, T. B. Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science. 343 (6173), 1244624(2014).
- Mattes, B., et al. Wnt/PCP controls spreading of Wnt/β-catenin signals by cytonemes in vertebrates. eLife. 7, 36953(2018).
- Bodeen, W. J., Marada, S., Truong, A., Ogden, S. K. A fixation method to preserve cultured cell cytonemes facilitates mechanistic interrogation of morphogen transport. Development. 144 (19), 3612-3624 (2017).
- Hall, E. T., Ogden, S. K. Preserve cultured cell cytonemes through a modified electron microscopy fixation. Bio-protocol. 8 (13), (2018).
- American Veterinary Medical Association. AVMA guidelines for the euthanasia of animals. American Veterinary Medical Association. , (2020).
- Harfe, B. D., et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 118 (4), 517-528 (2004).
- Chamberlain, C. E., Jeong, J., Guo, C., Allen, B. L., McMahon, A. P. Notochord-derived Shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. Development. 135 (6), 1097-1106 (2008).
- Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L., Luo, L. A global double-fluorescent Cre reporter mouse. Genesis. 45 (9), 593-605 (2007).
- Bacallao, R., Sohrab, S., Phillips, C. Handbook Of Biological Confocal Microscopy. Pawley, J. B. , Springer US. 368-380 (2006).
- Tagliaferro, P., Tandler, C. J., Ramos, A. J., Pecci Saavedra, J., Brusco, A. Immunofluorescence and glutaraldehyde fixation. A new procedure based on the Schiff-quenching method. Journal of Neuroscience Methods. 77 (2), 191-197 (1997).
- Neckel, P. H., Mattheus, U., Hirt, B., Just, L., Mack, A. F. Large-scale tissue clearing (PACT): Technical evaluation and new perspectives in immunofluorescence, histology, and ultrastructure. Scientific Reports. 6 (1), 1-13 (2016).
- Clendenon, S. G., Young, P. A., Ferkowicz, M., Phillips, C., Dunn, K. W. Deep tissue fluorescent imaging in scattering specimens using confocal microscopy. Microscopy and Microanalysis. 17 (4), 614-617 (2011).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved