Method Article
Generation of Patient-Derived Podocytes from Skin Biopsies
In This Article
Summary
This manuscript describes a two-step protocol to generate patient-specific podocytes from dermal fibroblasts via episomal reprogramming into human-induced pluripotent stem cells (hiPSCs) and subsequent differentiation into podocytes.
Abstract
Podocytes are epithelial cells sitting on the urinary site of the glomerular filtration barrier that contribute to the selective filter function of the glomerulus. Mutations in podocyte-specific genes can cause focal segmental glomerulosclerosis (FSGS), and podocytes are also affected in many other primary and secondary nephropathies. Due to their differentiated nature, primary cell culture models are limited for podocytes. Therefore, commonly conditionally immortalized cells are used. However, these conditionally immortalized podocytes (ciPodocytes) have several limitations: the cells can dedifferentiate in culture, especially when they reach confluency, and several podocyte-specific markers are either only slightly or not expressed at all. This brings the use of ciPodocytes and their applicability for physiological, pathophysiological, and clinical reach into question. Here, we describe a protocol for the generation of human podocytes-including patient-specific podocytes-from a skin punch biopsy by episomal reprogramming of dermal fibroblasts into hiPSCs and subsequent differentiation into podocytes. These podocytes resemble in vivo podocytes much better in terms of morphological characteristics, like the development of foot processes and the expression of the podocyte-specific marker. Finally, yet importantly, these cells maintain patients' mutations, resulting in an improved ex vivo model to study podocyte diseases and potential therapeutic substances in an individualized approach.
Introduction
Podocytes are specialized, post-mitotic renal epithelial cells that form the glomerular filtration barrier of the kidney along with the glomerular basement membrane (GBM), glomerular endothelial cells, and glycocalyx. Phenotypically, podocytes consist of a cell body and primary, microtubule-driven membrane extensions, as well as secondary extensions called foot processes1,2. The glomerular filtration barrier that filters urine from the blood is built of fenestrated endothelium, GBM, and a specialized type of intercellular junction that connects neighboring podocyte foot processes, called the slit diaphragm of podoctyes3. Under healthy conditions, proteins larger than albumin are retained from the filtration barrier due to their size and charge4.
Mutations in cytoskeletal- or podocyte-specific genes, as well as circulating factors affecting podocyte signaling pathways, are known to induce podocyte effacement, detachment, or apoptosis, resulting in proteinuria and glomerular sclerosis. In particular, cytoskeletal rearrangement, changes in podocyte polarity or damage of foot processes with an associated loss of slit junctions are pivotal5. Due to their terminally differentiated status, podocytes can hardly be replaced after detachment of the GBM. However, if podocytes are attached to the GBM, they can still recover from effacement and reform interdigitating foot processes6,7,8. Further understanding of the events that lead to podocyte damage in various glomerular disorders may provide novel therapeutic targets that will aid in developing treatments for these diseases. Podocyte damage is a hallmark of different glomerular diseases, including focal segmental glomerulosclerosis (FSGS), diabetic nephropathy, minimal change disease, and membranous glomerulonephropathy, requiring reliable podocyte ex vivo models to study the pathological mechanisms and potential treatment approaches for these diseases9,10. Podocytes can be studied ex vivo by classical primary cell culture based on the isolation of glomeruli by differential sieving11. However, due to the terminally differentiated state with limited proliferation capacity, most researchers use mouse or human ciPodocyte cell lines that express temperature-sensitive variants of the SV40 large T antigen. Alternatively, ciPodocytes are isolated from transgenic mice harboring the SV40 Tag immortalizing gene1,12.
CiPodocytes proliferate at 33 °C, but enter growth arrest and start differentiating at 37 °C13,14. It has to be kept in mind that experimental data obtained with these cells should be interpreted with certain caution, as the cells are generated using an unnatural gene insertion15. As these cells harbor an immortalizing gene, the cellular physiology is altered because of ongoing proliferation12. Podocyte cell lines generated by this approach have recently been questioned, as mouse, human, and rat ciPodocytes express less than 5% of synaptopodin and nephrin at the protein level, as well as NPHS1 and NPHS2 at the mRNA level compared to glomerular expression16. Moreover, most podocyte cell lines do not express nephrin17,18. Chittiprol et al. also described a significant difference in cell motility and responses to puromycin and doxorubicin in ciPodocytes16. Podocytes can be found in urine after detachment from the GBM in different glomerular diseases19,20,21,22. Viable urinary podocytes can be cultivated ex vivo for up to 2-3 weeks, but most cells undergo apoptosis23,24. Interestingly, podocytes are not only found in the urine of patients with glomerular disease but also in the urine of healthy subjects, most likely when they are senescent-again with a limited potential of replication in culture24. Furthermore, the urinary-derived podocyte number is limited, and the cells dedifferentiate in culture, show less foot processes, change morphology, and most importantly have limited proliferation capacity. The expression of podocyte-specific genes is absent, disappears within a few weeks, or varies among these cell clones. Some cells positive for the podocyte-specific marker co-expressed the marker of tubular epithelial cells or myofibroblasts and mesangial cells, which suggests dedifferentiation and/or transdifferentiation of the cultured urinary podocytes24,25.
Recently, the generation of ciPodocyte cell lines derived from the urine of patients and healthy volunteers by transduction with a thermo-sensitive SV40 large T antigen and hTERT has been described26. mRNA expression for synaptopodin, nestin, and CD2-associated protein was detected, but podocin mRNA was absent in all clones. In addition to the problems with urinary podocytes, these cells also contain the inserted immortalizing gene, resulting in the disadvantages discussed above.
In contrast, human induced pluripotent stem cells (hiPSCs) have a huge capacity to self-renew and differentiate into multiple cell types under appropriate conditions. It has been shown before that hiPSCs can serve as an almost unlimited source of podocytes27,28.
Here, a two-step protocol to generate patient-specific podocytes from dermal fibroblasts of skin punch biopsies with subsequent episomal reprogramming into hiPSCs and a final differentiation into hiPSC-derived podocytes is described (Figure 1).
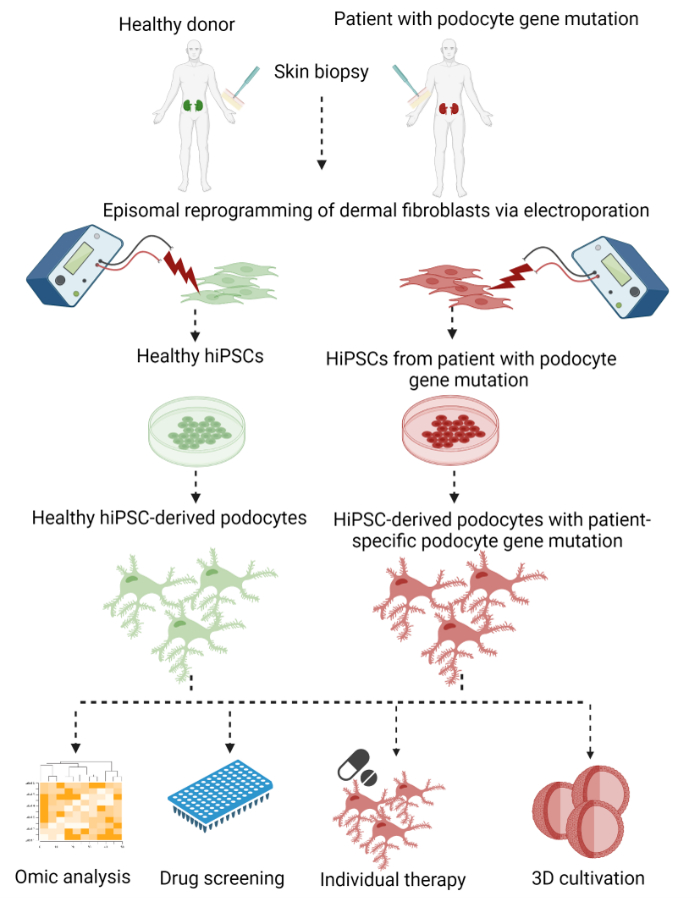
Figure 1: Protocol to generate patient-specific hiPSC-derived podocytes. Graphical overview of the protocol to generate patient-specific podocytes from dermal fibroblasts of a skin biopsy by reprogramming into hiPSCs and differentiation into podocytes. Please click here to view a larger version of this figure.
As a first step, somatic dermal fibroblasts were outgrown from a skin punch biopsy and reprogrammed into hiPSCs using an integration-free method by electroporation with plasmids expressing the transcription factors OCT3/4, KLF4, SOX2, and c-MYC29,30,31. Arising hiPSC colonies were subsequently selected and expanded. Differentiation began with the induction of the mesodermal lineage by activation of the WNT signaling pathway, followed by the generation of nephron progenitor cells that were still able to proliferate. Finally, the cells were differentiated into podocytes. In this procedure, we modified and combined previously published protocols for episomal reprogramming for the generation of hiPSCs by Bang et al.32 and Okita et al.33, as well as a protocol for the differentiation of hiPSCs into podocytes by Musah et al.28,34,35.
Indeed, podocytes generated by our protocol had a phenotype closer to podocytes in vivo, regarding the development of a distinct network of primary and secondary foot processes and the expression of podocyte-specific markers, like synaptopodin, podocin, and nephrin. With the use of hiPSC-derived podocytes, the patient's genetic background is maintained during reprogramming and differentiation. This enables patient-specific podocyte disease modelling and the discovery of potential therapeutic substances ex vivo in an almost unlimited cell number. Moreover, this protocol is minimally invasive, cost-efficient, ethically acceptable and may facilitate new avenues for drug development.
Protocol
The protocol was approved by the ethics committee of Friedrich-Alexander University Erlangen-Nuremberg (251_18B), and all patients and probands gave written consent. All experiments were performed in accordance with relevant guidelines and regulations. For composition of all media and solutions used here see Table 1.
1. Outgrowth of dermal fibroblasts from a skin punch biopsy
- Transfer the skin punch biopsy to a sterile 15 mL conical tube with 10 mL of prewarmed fibroblast medium.
- Remove the medium and wash the skin punch biopsy three times with 5 mL of sterile, prewarmed 1x phosphate-buffered saline (PBS). Transfer the skin punch biopsy with sterile forceps to a 10 cm cell culture plate and cut it widthwise in three or four pieces with a sterile scalpel, leaving the epidermis and dermis.
- Transfer each piece to a sterile 35 mm plastic cell culture dish and press it gently to the culture dish. Remove excess medium around the biopsy pieces. Allow to dry for 5-10 min until the liquid evaporates and the biopsy is attached to the cell culture plastic.
- Add 1 mL of fibroblast medium dropwise with a 1,000 µL pipette around the biopsy and fill up carefully to a final volume of 3 mL. Cultivate at 37 °C, 5% CO2 for 7 days without feeding and moving the dish. Carefully change the medium to fresh, prewarmed fibroblast medium. After 7 days, outgrown spindle-like fibroblasts can be monitored around the skin biopsy using a phase contrast microscope36.
NOTE: When outgrown fibroblasts reach the rim of the dish, proceed again from step 1.3 to generate another batch of fibroblasts. - For expansion of the outgrown fibroblasts, wash with 2 mL of prewarmed 1x PBS, dissociate the fibroblasts with 1 mL of 1x trypsin-ethylenediaminetetraacetic acid (EDTA) and incubate for 5 min at 37 °C, 5% CO2. Transfer the detached cells to a 15 mL conical tube and wash the cell culture dish with 2 mL of fresh, prewarmed fibroblast medium.
- Pool the washing medium in the tube to neutralize the dissociation agent and centrifuge at 200 x g for 5 min at 20 °C. Aspirate the supernatant and resuspend the cell pellet with 1 mL of fresh, prewarmed fibroblast medium. Count the cells with a Neubauer chamber or automated cell counter using trypan blue and seed 2.5 x 103 to 5 x 103 fibroblasts per cm2 in fresh cell culture flasks.
- Place the flasks back in the incubator and distribute the cells evenly by moving the flasks three times in each direction. The next day, replace the medium with fresh, prewarmed fibroblast medium. Change the medium twice a week and split when the fibroblasts reach around 80% confluency.
2. Freezing dermal fibroblasts
- To freeze the fibroblasts, wash them with 5 mL of prewarmed 1x PBS. Aspirate the PBS and dissociate the fibroblasts with 4 mL of 1x trypsin-EDTA. Place the flask back in the incubator and incubate for 5-7 min at 37 °C at 5% CO2. Monitor detachment using a phase contrast microscope and tap against the side of the flask to detach the cells.
- Transfer the detached cells to a 15 mL conical tube. Wash the cell culture flask with 5 mL of fresh, prewarmed fibroblast medium. Pool the washing medium in the conical tube and centrifuge at 200 x g for 5 min at 20 °C.
- Aspirate the supernatant and resuspend the cell pellet with 2 mL of fresh, prewarmed fibroblast medium. Count the cells and transfer 1 x 106 cells to a new conical tube. Centrifuge at 200 x g for 5 min at 20 °C.
- Aspirate the supernatant and resuspend the cell pellet with 1 mL of cold fibroblast freezing medium consisting of 90% fetal calf serum and 10% dimethyl sulfoxide (DMSO). Transfer to a cryovial, place in a freezing container, and freeze overnight at -80 °C. For long term storage, place the cryovial in a liquid nitrogen tank the next day.
3. Episomal reprogramming of fibroblasts to generate hiPSCs
- For episomal reprogramming, use 1.5 x 106 fibroblasts from passage 4 to 8. To reach this cell number, use two to three 250 mL flasks with around 70% confluency.
- The day before reprogramming, split the fibroblasts in a 1:2 ratio to ensure their proliferative state. Therefore, aspirate the medium and wash the flasks with 5 mL of prewarmed 1x PBS per flask. Aspirate the PBS and dissociate the fibroblasts with 4 mL of 1x trypsin-EDTA. Place the flasks back in the incubator and incubate for 5-7 min at 37 °C and 5% CO2. Monitor detachment using a phase contrast microscope and tap against the side of the flask to detach the cells from the plastic surface.
- Transfer the detached cells to a 50 mL conical tube and wash the cell culture flasks with 5 mL of fresh, prewarmed fibroblast medium. Pool the washing medium in the conical tube and centrifuge at 200 x g for 5 min at 20 °C. Aspirate the supernatant and resuspend the cell pellet in 6 mL of fresh, prewarmed fibroblast medium.
- Prepare six new cell culture flasks by adding 5 mL of fresh, prewarmed fibroblast medium per flask. Add 1 mL of fibroblast cell suspension to each flask. Place the flasks back in the incubator and distribute the cells evenly by moving the flasks three times in each direction.
- For episomal reprogramming, coat two 10 cm cell culture plates suitable for hiPSC culture with 4 mL of cold coating solution per plate. Incubate the plates for 1 h at 37 °C.
NOTE: To culture hiPSCs, different cell culture plastics and additional extracellular matrix coating are required (see Table of Materials). - Remove the media from the fibroblasts and wash with 10 mL of prewarmed 1x PBS per flask. Aspirate the PBS and dissociate the fibroblasts with 4 mL of 1x trypsin-EDTA per flask by incubating for 5-7 min at 37 °C. Monitor detachment using a phase contrast microscope and, if necessary, tap against the side of the flask to detach the cells from the plastic surface.
- Transfer the detached cells to a 50 mL conical tube. Wash the empty flasks with 6 mL of prewarmed fibroblast medium to collect the remaining cells and pool in the conical tube. Centrifuge at 200 x g for 5 min at 20 °C. Aspirate the supernatant and resuspend the cell pellet in 3 mL of fresh, prewarmed 1x PBS.
- Count the cells and transfer 1.5 x 106 fibroblasts in a new conical tube. Centrifuge at 200 x g for 5 min at 20 °C. Discard the supernatant and resuspend the cell pellet in 5 mL of electroporation medium and centrifuge again. In the meantime, aspirate the coating solution from the coated 10 cm cell culture plates and add 7 mL of prewarmed fibroblast medium.
- After centrifugation, discard the supernatant and resuspend the cell pellet in electroporation medium with a concentration of 1.5 x 106 cells in 250 µL. Transfer the 250 µL cell suspension to an electroporation cuvette with a 4 mm gap distance (see Table of Materials).
- Prepare a plasmid transfection mix by adding 4 µg of each plasmid (pCXLE-hOCT3/4, pCXLE-hSK, pCXLE-hMLN) to a total volume of 50 µL of electroporation medium. Transfer to the cuvette and mix by flicking gently. Electroporate with one pulse at 280 V.
- Cut a pipette tip using sterile scissors and transfer 125 µL of the electroporated fibroblasts to each of the prepared 10 cm cell culture plates. Distribute the cells by agitating the plates three times in all directions and place them back in the incubator. Incubate overnight at 37 °C with 5% CO2 without disturbance.
- To remove dead cells, replace the medium the next day with 7 mL of fresh, prewarmed fibroblast medium. Change the medium to hiPSC culture medium 2 days after electroporation and replace it every other day for the next 20 days.
4. Selection, expansion, and quality control of generated hiPSCs
- Monitor the cells daily for at least 20 days using a phase contrast microscope with a 10x or 20x objective to observe the formation of hiPSC colonies after electroporation. If hiPSC colonies are around 300 µm diameter in size with distinct borders and the hiPSCs show a high nuclei-to-body ratio, the hiPSC colonies are ready for selection by picking (Figure 2B,C and Figure 3).
- Prior to picking, coat a 96-well plate suitable for hiPSC culture with 100 µL of coating solution per well and incubate for 1 h at 37 °C. During incubation, mark colonies of interest at the bottom of the cell culture dish with a pen. For final preparation of the 96-well plate, remove the coating solution and add 100 µL of prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632 to each well.
- Wash the cells with prewarmed 1x PBS and add fresh, prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632 before picking to remove dead cells. To pick hiPSC colonies, use a gauge needle and divide the hiPSC colonies into small pieces by drawing a grid into each colony.
- Using a phase contrast microscope, check that the colonies are successfully divided into pieces. Transfer them with a 100 µL pipette to the prepared 96-well plate. Hold the pipette upright over the colony without touching the cells to avoid scratching and loss of the colony.
- Place the 96-well plate in the incubator at 37 °C with 5% CO2 and let the cells attach overnight without disturbance. Keep the dishes with the fibroblast and hiPSC mix, in case picking is not successful. Therefore, remove ROCK inhibitor Y27632 by changing the medium to hiPSC culture medium and place the plates back in the incubator.
- The next day, change medium of the 96-well plate to 200 µL of fresh hiPSC culture medium and change it every other day. Monitor the 96-well plate the next day and mark the wells with successfully picked clones.
NOTE: If the picked hiPSC clones are not fully selected after the first try and the fibroblasts can be found in the well, repeat picking from the 96-well or scrape the fibroblasts off the plate.
5. Expansion of selected hiPSC clones
- Monitor the hiPSC clones using a phase contrast microscope. If the selected hiPSCs reach around 70% confluency, the hiPSC clones are ready for expansion.
- Coat a 48-well plate suitable for hiPSC culture with 250 µL of coating solution per well for 1 h at 37 °C. Replace the coating solution with 200 µL of fresh, prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632.
- To detach the hiPSCs from the 96-well plate, scrape the plastic surface with a 1,000 µL pipette tip. Transfer the detached hiPSCs from the 96-well plate to a 48-well plate. Change the medium the next day to fresh, prewarmed hiPSC culture medium to remove dead cells and the ROCK inhibitor Y27632.
- If the hiPSC clones reach around 70% confluency, transfer the hiPSCs into a 24-well plate. Therefore, coat a 24-well plate suitable for hiPSC culture with 400 µL of coating solution per well for 1 h at 37 °C. Replace the coating solution with 400 µL of fresh, prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632. Aspirate the medium from the 48 wells and wash with 500 µL of prewarmed 1x PBS.
- Replace the PBS with 100 µL of enzymatic cell detachment solution containing 10 µM ROCK inhibitor Y27632 and incubate at 37 °C for 4 min. Rinse the hiPSCs off the plate using a 1,000 µL pipette. Transfer the dissociated cells to a 15 mL conical tube. Wash the empty 48-well plate with hiPSC culture medium containing 10 µM ROCK inhibitor Y27632 and pool in the conical tube.
- Centrifuge at 200 x g for 5 min at 20 °C. Aspirate the supernatant and resuspend the hiPSCs in 1 mL of hiPSC culture medium containing 10 µM ROCK inhibitor Y27632 and transfer to a 24-well plate. Place the plate in the incubator at 37 °C at 5% CO2 and distribute the cells by agitating the plate three times in all directions. Let the cells attach overnight without disturbance.
- The next day, change the medium to hiPSC culture medium without ROCK inhibitor Y27632.
NOTE: If hiPSC clones reach around 70% confluency, transfer the hiPSCs to a 12-well plate by repeating steps 5.4 to 5.7 with increased volumes suitable for a 12-well format. If the hiPSCs from the 12-well plate reach around 70% confluency, transfer the hiPSC clones to a 6-well plate with increased volumes for a 6-well format.
6. Freezing of selected hiPSC clones
- To freeze selected hiPSC clones, aspirate the medium from a 6-well plate. Wash the wells with 2 mL of 1x PBS. Aspirate and dissociate the hiPSCs with 1 mL of enzymatic cell detachment solution containing 10 µM Y27632. Place the plate back in the incubator and incubate for 4 min at 37 °C and 5% CO2.
- Rinse the hiPSCs off the plate using a 1,000 µL pipette and transfer the detached cells to a 15 mL conical tube. Wash the plate with 2 mL of fresh, prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632. Pool in the conical tube and centrifuge at 200 x g for 5 min at 20 °C. Aspirate the supernatant and resuspend the cell pellet with 2 mL of fresh, prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632.
- Count the cells and transfer 1 x 106 cells to a new conical tube. Centrifuge at 200 x g for 5 min at 20 °C. Aspirate the supernatant, resuspend the cell pellet in 1 mL of cold, serum-free hiPSC freezing medium, and freeze in a cryovial using a freezing container overnight at -80 °C. For long-term storage, place the cryovial in a liquid nitrogen tank the next day.
7. HiPSC quality control
- Characterization of hiPSC morphology
- Check the characteristic hiPSC morphology using a phase contrast microscope with a 20x objective. After reprogramming, the cell morphology changes from long, spindle-like fibroblasts to small, round hiPSCs, presenting a high nuclei-to-body ratio growing in colonies with distinct borders (Figure 2C).
- If the hiPSC colonies grow too dense and spontaneous differentiation occurs, scrape the differentiated parts off the plate using a pipette tip (Figure 3). Since hiPSCs have a high proliferation rate, feed the hiPSC cultures every day with fresh, prewarmed hiPSC feeding medium.
- Characterization of pluripotency markers by immunofluorescence staining
- To check the generated hiPSCs for pluripotency marker via immunofluorescence staining, place plastic coverslips in a 24-well plate and coat with 250 µL of coating solution for 1 h at 37 °C and 5% CO2. Replace the coating solution with 1 mL of prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632.
- Seed 1.9 x 104 dissociated hiPSCs per 24-well. Let the hiPSCs attach overnight at 37 °C and 5% CO2 without disturbance. See steps 5.4 to 5.6 for dissociation of the hiPSCs. Replace the medium the next day with hiPSC culture medium without ROCK inhibitor Y27632.
- For staining, wash the cells with 1 mL of prewarmed 1x PBS and subsequently fix the hiPSCs with 4% paraformaldehyde for 10 min at room temperature. Wash the fixed hiPSCs with 1x PBS and block unspecific binding sites with 200 µL of preincubation solution per well for 1 h at room temperature. Dilute primary antibodies Ki67 (1:300), OCT3/4 (1:200), SSEA4 (1:100) in antibody diluent.
- Clean a plastic foil with 70% ethanol and place it in a staining chamber filled with a water reservoir. Place droplets of 30 µL primary antibody dilutions on the foil. Place the fixed samples upside down in the dilution and incubate overnight at 4 °C.
- The next day, wash the samples three times with 1x PBS for 5 min and place 30 µL droplets of secondary antibody dilutions (1:1,000) on clean spots on the foil. Place the cover slides upside down in the dilution. Incubate for 1 h at room temperature in the dark.
- After incubation, wash three times with 1x PBS for 5 min. Mount the samples on glass slides in the mounting medium containing DAPI. Allow to dry overnight at room temperature and in the dark and image the samples using a confocal microscope (Figure 4).
- Exclusion of bacteria and mycoplasma contamination
- To exclude bacteria contamination, transfer 500 µL of dissociated hiPSCs to 5 mL of prewarmed Luria-Bertani (LB) medium. See steps 5.4 to 5.6 for dissociation of the hiPSCs. Incubate overnight at 37 °C. If the LB medium appears turbid, bacteria contamination has likely occurred. Discard the cells.
- To exclude mycoplasma contamination, collect medium that has not been replaced for 2 days from 6 wells with around 90% confluent hiPSCs. Use quantitative polymerase chain reaction (qPCR) to detect mycoplasma contamination. Many commercial mycoplasma detection kits are available for this purpose.
8. Differentiation of hiPSCs into podocytes
- Preparation and storage of growth factors
- To prepare a stock concentration of 10 mM Y27632, reconstitute 10 mg of Y27632 (molecular weight of 320.26 g/mol) in 3.12 mL of sterile water. Store 100 µL aliquots at -20 °C and dilute 1:1,000 in cell culture medium to reach a final concentration of 10 µM.
- To prepare a stock concentration of 30 mM CHIR99021, reconstitute 5 mg of CHIR99021 (molecular weight of 465.34 g/mol) in 358.2 µL of sterile DMSO. Store 50 µL aliquots at -20 °C and dilute 1:10,000 in cell culture medium to reach a final concentration of 3 µM.
- To prepare a stock concentration of 100 µg/mL activin A, reconstitute 100 µg of activin A in 1 mL of 1x PBS containing 0.1% bovine serum albumin (BSA). Store 100 µL aliquots at -20 °C and dilute 1:2,000 in cell culture medium to reach a final concentration of 50 ng/mL.
- To prepare a stock concentration of 100 µg/mL bone morphogenic protein 7 (BMP7), reconstitute 100 µg of BMP7 in 1 mL of sterile water containing 0.1% BSA. Store 100 µL aliquots at -20 °C and dilute 1:2,000 in cell culture medium to reach a final concentration of 50 ng/mL.
- To prepare a stock concentration of 100 µg/mL vascular endothelial growth factor (VEGF), reconstitute 100 µg of VEGF in 1 mL of sterile water. Store 100 µL aliquots at -20 °C and dilute 1:4,000 in cell culture medium to reach a final concentration of 25 ng/mL.
- To prepare a 10 mM stock concentration of all-trans retinoic acid, reconstitute 10 mg of all-trans retinoic acid in 3.33 mL of sterile DMSO. Store 100 µL aliquots at -20 °C and dilute 1:100 in cell culture medium to reach a final concentration of 0.5 µM.
- Activation of WNT signaling pathway to induce mesoderm lineage
- For differentiation of hiPSCs into hiPSC-derived podocytes, coat cell culture plates or flasks suitable for hiPSC culture with coating solution for 1 h at 37 °C and 5% CO2. The total volume of coating solution is 1 mL per 6-well culture plate or 4 mL per 10 cm cell culture plate.
- Aspirate the medium and wash the hiPSCs with prewarmed 1x PBS. Aspirate the PBS and add enzymatic cell detachment solution containing 10 µM Y27632 at a total volume of 1 mL per 6-well culture plate or 4 mL per 10 cm cell culture plate.
- Place the plate back in the incubator and incubate for 4 min at 37 °C and 5% CO2. Rinse the hiPSCs off the plate using a 1,000 µL pipette. Transfer the detached cells to a conical tube.
- Wash the plate with fresh, prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632. Pool the washing medium in the conical tube to neutralize the dissociation reagent. Centrifuge at 200 x g for 5 min at 20 °C. Aspirate the supernatant and resuspend the cell pellet with 2 mL of fresh, prewarmed hiPSC culture medium containing 10 µM ROCK inhibitor Y27632.
- Count the cells and seed 1 x 104 hiPSCs/cm2. Distribute the cells by agitating three times in all directions. Let the hiPSCs attach overnight at 37 °C with 5% CO2 without disturbance.
- The next day, replace the medium with 2 mL of prewarmed mesoderm differentiation medium containing 1x B27 supplement, 1% penicillin-streptomycin, 3 µM CHIR99021, 50 ng/mL activin A and 10 µM ROCK inhibitor Y27632.
- Differentiation into nephron progenitor cells
- After 2 days, change the mesoderm medium to 2 mL of prewarmed nephron progenitor differentiation medium containing 1x B27 supplement, 1% penicillin-streptomycin, 3 µM CHIR99021, 50 ng/mL activin A, and 50 ng/mL BMP7 per 6-well culture plate or 6 mL per 10 cm cell culture plate. Change the medium every other day for the next 14 days.
NOTE: Nephron progenitor cells proliferate and can be passaged and frozen after 7 days of differentiation in nephron progenitor expansion medium. - To split the nephron progenitor cells, coat the cell culture plastics suitable for hiPSC culture with coating solution for 1 h at 37 °C. Replace the coating solution with nephron progenitor expansion medium. Aspirate the media and wash the cells with prewarmed 1x PBS. Remove the PBS and dissociate the cells with 1x trypsin-EDTA.
- Incubate for 5 min at 37 °C and 5% CO2. Monitor detachment of the cells using a phase contrast microscope. If necessary, tap against the side of the plastic to detach the cells from the surface. Rinse the cells off the plate and transfer to a conical tube. Wash the empty flask or plate with the same volume of prewarmed nephron progenitor expansion medium as the dissociation reagent.
- Pool the washing medium in the conical tube. Centrifuge at 200 x g for 5 min at 20 °C. Aspirate the supernatant and resuspend the cell pellet with 2 mL of fresh, prewarmed nephron progenitor expansion medium. Count the cells and seed 1.5 x 104 nephron progenitor cells per cm2 for further differentiation.
NOTE: To freeze the nephron progenitor cells, resuspend 1 x 106 cells in 1 mL of cold serum-free cryopreservation medium and transfer to a cryovial. Freeze in a freezing container at -80 °C overnight and transfer to a liquid nitrogen tank for long-term storage. - Place the plates back in the incubator and distribute the cells evenly by agitation three times in all directions. Change the medium the next day to fresh, prewarmed nephron progenitor differentiation medium. Replace the medium every other day until the cells are differentiated for 14 days in nephron progenitor differentiation medium.
- After 2 days, change the mesoderm medium to 2 mL of prewarmed nephron progenitor differentiation medium containing 1x B27 supplement, 1% penicillin-streptomycin, 3 µM CHIR99021, 50 ng/mL activin A, and 50 ng/mL BMP7 per 6-well culture plate or 6 mL per 10 cm cell culture plate. Change the medium every other day for the next 14 days.
- Terminal differentiation into hiPSC-derived podocytes
- Aspirate the medium and add 2 mL of prewarmed podocyte differentiation medium containing 1x B27 supplement, 1% penicillin-streptomycin, 3 µM CHIR99021, 50 ng/mL activin A, 50 ng/mL BMP7, 25 ng/µL VEGF, and 0.5 µM all-trans retinoic acid per 6-well culture plate or 6 mL per 10 cm cell culture plate. Place the plates back in the incubator at 37 °C and 5% CO2. Change the medium every other day for 4 days with fresh, prewarmed podocyte differentiation medium.
- To keep the hiPSC-podocytes in culture after differentiation, feed the cells twice a week with podocyte maintenance medium containing 10% fetal calf serum, 1% penicillin-streptomycin, and 0.1% insulin-transferrin-selenium.
NOTE: Terminal differentiated hiPSC-podocytes do not proliferate anymore. However, it is possible to keep them in cell culture for up to 4 weeks.
- Thawing and expansion of frozen nephron progenitor cells
- Coat a new cell culture plastic flask suitable for hiPSC culture with coating solution for 1 h at 37 °C and 5% CO2. Replace the coating solution with 5 mL of nephron progenitor expansion medium.
- Prepare a 15 mL conical tube with 7 mL of prewarmed nephron progenitor expansion medium. Thaw the frozen cells at 37 °C in a water bath without movement for approximately 20 s. Take the cryovial out of the water bath when half of the cells are thawed, and some ice is remaining.
- Spray the cryovial with 70% ethanol and place in a biosafety cabinet. Transfer thawed cells to the conical tube with the prewarmed nephron progenitor expansion medium. Use a 1,000 µL pipette and let the cells run down the wall of the tube very slowly.
- Wash the empty cryovial with 1 mL of nephron progenitor expansion medium to collect the remaining cells and pool in the conical tube. Centrifuge at 200 x g for 5 min at 20 °C and resuspend the cell pellet in fresh, prewarmed nephron progenitor expansion medium.
- Transfer the thawed cells to the coated flask and change the medium the next day to fresh, prewarmed nephron progenitor expansion medium. Before further differentiation, let the cells proliferate in nephron progenitor expansion medium by changing the medium every other day, and proceed from step 8.3.5.
9. Characterization of hiPSC-derived podocytes by immunofluorescence staining
- To check the differentiated podocytes for the podocyte-specific marker via immunofluorescence staining, place plastic coverslips in a 24-well plate and coat with 250 µL of coating solution for 1 h at 37 °C and 5% CO2. Replace the coating solution with 1 mL of prewarmed nephron progenitor expansion medium. Seed 2.5 x 104 dissociated nephron progenitor cells per 24-well plate. See steps 8.3.2 to 8.3.4 for dissociation of nephron progenitor cells.
- Let the cells attach overnight at 37 °C and 5% CO2 without disturbance. Replace the medium the next day with prewarmed nephron progenitor differentiation medium.
- Finish differentiation by changing the medium every other day until the cells are differentiated for a total of 14 days in nephron progenitor differentiation medium and an additional 5 days in podocyte differentiation medium.
- After the final differentiation, wash the cells with 1 mL of prewarmed 1x PBS. Fix the hiPSC-podocytes with 4% paraformaldehyde for 10 min at room temperature. Wash the fixed cells with 1x PBS and block unspecific binding sites with 200 µL of preincubation solution per well for 1 h at room temperature.
- Dilute the primary antibodies synaptopodin (1:200), podocin (1:100), and nephrin (1:25) in antibody diluent and place droplets of 30 µL primary antibody dilution on a clean plastic foil in a staining chamber containing a water reservoir.
- Place cover slides with fixed hiPSC-podocytes upside down in the dilution. Incubate overnight at 4 °C. The next day, wash three times with 1x PBS for 5 min. Dilute the secondary antibodies (1:1,000) in 1x PBS and place droplets of 30 µL secondary antibody dilution on clean spots on the foil. Place the cover slides upside down in the dilution.
- Incubate for 1 h at room temperature in the dark. After incubation, wash three times with 1x PBS for 5 min. Mount the samples on glass slides in mounting medium containing DAPI. Allow to dry overnight at room temperature and in the dark. Image the samples using a confocal microscope.
Results
With this step-by-step protocol that combines episomal reprogramming and differentiation, it is possible to generate podocytes carrying a patient's mutation in a podocyte-relevant gene. This enables the analysis of disease-specific podocyte alterations ex vivo. The patient's genetic background is preserved during the protocol during all the different cell stages. In addition, the limitation of insufficient cell number of terminally differentiated non-proliferating podocytes can be overcome by using hiPSC-derived podocytes. Although it takes several months until hiPSC-podocytes are generated from outgrown dermal fibroblasts via reprogramming into hiPSCs and subsequent differentiation into podocytes, it is possible to freeze cells at three different steps of the protocol (Figure 2). Freezing is possible for fibroblasts, hiPSCs, and nephron progenitor cells after differentiation in nephron progenitor differentiation medium for 7 days. Therefore, the generation of working cell banks and large-scale experiments is possible.
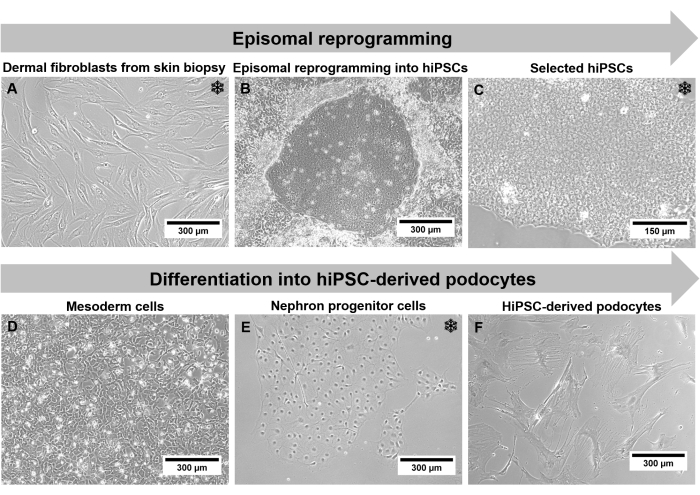
Figure 2: Individual steps of the protocol through a phase contrast microscope. (A) Outgrown dermal fibroblasts. (B) hiPSC colony formation after reprogramming. (C) Selected hiPSC culture. (D) Mesoderm cells after 2 days of differentiation. (E) Nephron progenitor cells after 14 days of differentiation in nephron progenitor differentiation medium. (F) Terminal differentiated hiPSC-derived podocytes. Scale bars represent 300 µm (A,B,D-F) and 150 µm (C). The snowflake highlights possible freezing steps. Please click here to view a larger version of this figure.
As the generated hiPSC-derived podocytes traverse a long protocol with drastic changes regarding cell type and morphology, cell characterization is obligatory. Cell morphology, like size and cell shape, as well as growth behavior, can be monitored using a phase contrast microscope. Fibroblasts present a long spindle-like phenotype with a size of 150 µm to 300 µm (Figure 2A). After reprogramming, colonies with 50 µm small hiPSCs occur. These colonies display distinct borders and hiPSCs distinguished with a high nuclei-to-body ratio and increased proliferation rate (Figure 2C). Since podocytes are terminally differentiated cells, the proliferation rate decreases with progressive differentiation, and the cell morphology changes to 300 µm large star-shaped podocytes with prominent foot processes (Figure 2F).
Confluency is a critical point of hiPSC culture, and spontaneous differentiation can appear when colonies grow too dense (Figure 3). These colonies must be removed before passaging by scraping the affected parts of the culture dish.
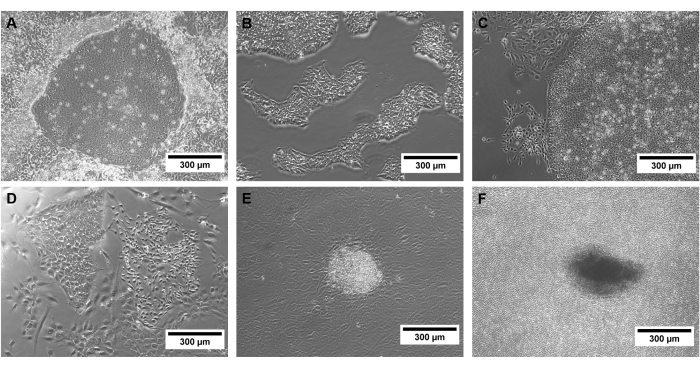
Figure 3: Examples of hiPSCs of different quality. Phase contrast images of successfully (A) reprogrammed hiPSC colonies and (B) selected hiPSCs, as well as (C) spontaneous differentiation, (D,E) non-hiPSC colonies, and (F) a too-dense hiPSC culture. Scale bars represent 300 µm. Please click here to view a larger version of this figure.
The different cell types of this protocol must be validated regarding the expression of a specific marker. After reprogramming, hiPSCs regain pluripotency capacity and express proliferation and pluripotency markers like SSEA4, OCT3/4, and Ki67 (Figure 4)38,39,40. It is known that reprogramming, as well as the lengthy culture of hiPSCs and differentiation, can lead to an abnormal karyotype, or rather induce mutations41. Therefore, the genetic background of the cultured cells should be monitored over time and at different passages by g-banding and whole exome sequencing.
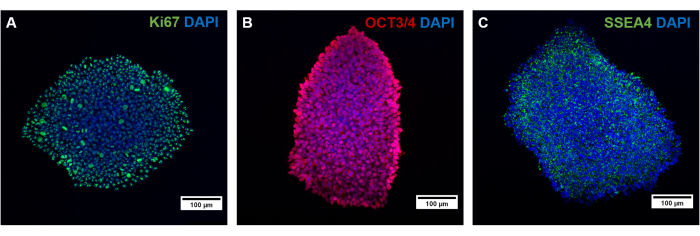
Figure 4: Characterization of generated hiPSCs by immunofluorescence staining. Characterization of generated hiPSCs by staining for (A) the proliferative marker Ki67, as well as the pluripotency markers (B) OCT3/4 and (C) SSEA4. Scale bars represent 100 µm. Please click here to view a larger version of this figure.
Morphologically, hiPSC-derived podocytes appear star-shaped and express a distinct network of long primary and secondary foot processes compared to ciPodocytes (Figure 5). HiPSC-derived podocytes express podocyte-specific marker proteins like synaptopodin, nephrin, and podocin (Figure 6A-C)42.
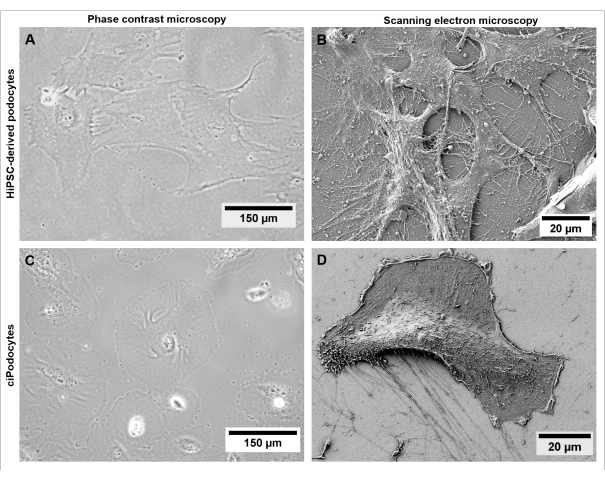
Figure 5: Morphology of hiPSC-derived podocytes compared to ciPodocytes. Comparison of (A,B) hiPSC-derived podocytes from a healthy donor to (C,D) ciPodocytes regarding cell morphology and filopodia by using a phase contrast microscope (A,C) and scanning electron microscope (B,D). Scale bars represent 150 µm (A,C) and 20 µm (B,D). Please click here to view a larger version of this figure.
Therefore, the comparison of hiPSC-derived podocytes not only from healthy donors, but also from patients with mutations in podocyte-specific genes, allows for individualized characterization of the patient's mutation ex vivo.
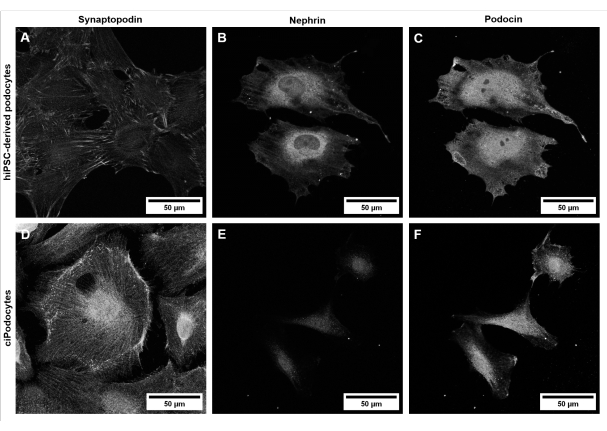
Figure 6: Characterization of a podocyte-specific marker in hiPSC-derived podocytes and ciPodocytes. Comparison of (A-C) hiPSC-derived podocytes from a healthy donor to (D-F) ciPodocytes regarding podocyte-specific marker proteins synaptopodin (A,D), nephrin (B,E), and podocin (C,F). Scale bars represent 50 µm. Please click here to view a larger version of this figure.
Table 1. Composition of all cell culture media and solutions used in the study. Please click here to download this Table.
Discussion
This cell culture-based protocol combines the episomal reprogramming of human dermal fibroblasts into patient-specific hiPSCs and subsequent differentiation into hiPSC-derived podocytes. This allows us to study mutation-related alterations of podocytes from patients with genetic glomerular disease regarding podocyte injury. The protocol to reprogram dermal fibroblasts with an integration-free method by electroporation is adapted from the published work of Bang et al.32 and Okita et al.33. The protocol to differentiate podocytes from hiPSCs is adapted from the published protocol from Musah et al.28,34,35. There are already publications available describing the generation of podocytes from hiPSCs27,34,35. However, the protocol provided here is optimized and less expensive regarding the differentiation of hiPSCs into podocytes. Compared to the published protocol from Musah et al., we tested this protocol on different coating reagents, like vitronectin, laminin silk-511, and solubilized basement membrane matrix. The concentrations of vitronectin and laminin silk-511 could be decreased to 2.5 µg/mL instead of 5 µg/mL28,34,35. Furthermore, it was possible to decrease the concentrations of BMP7 and activin A-two very expensive growth factors-by 50%, from 100 ng/mL to 50 ng/mL.
This enables less expensive differentiation. Nephron progenitor cells from day 7 were still proliferating, and the possibility of freezing was shown before. We expanded these cells after thawing and before final differentiation in basic medium containing Dulbecco's modified Eagle medium (DMEM) and B27 for several days, decreasing the costs even further. In addition to the differentiation steps, this protocol describes the outgrowth of fibroblasts from skin biopsies with subsequent generation of patient-specific hiPSCs via episomal reprogramming. The combination of these two methods enables the generation of patient-specific podocytes. Therefore, a complete step-by-step protocol to generate patient-specific hiPSC-derived podocytes is provided here that was not described before in such detail.
As the overall protocol includes several different cell types, it is crucial to characterize the generated cell types at different steps. Cells are in culture for an extended period of time, so quality control should be performed at different passages. When working with hiPSCs, daily feeding as well as cell behavior and morphology monitoring is necessary. Sterility of the differentiation media must be ensured by filter sterilization through a 0.2 µm filter. The whole protocol, from skin biopsy to hiPSC-derived podocytes, takes several months, but it is possible to freeze the cells at distinct stages of the process. Fibroblasts, selected hiPSC clones, and proliferative nephron progenitor cells after 7 days in nephron progenitor differentiation medium can be frozen, and a working cell bank can be generated.
Even though hiPSC-derived healthy podocytes develop a distinct network of primary and secondary foot processes (Figure 5A,B) and express typical podocyte-specific marker (Figure 6A-C), characteristic slit diaphragms, as seen in vivo, are hard to mimic in classical two-dimensional cell culture models. Moreover, intercell communication with other glomerular cell types is not possible in this mono-culture setting.
Due to their terminal differentiated state and lack of proliferation capacity, it is difficult to study podocytes ex vivo. With the help of conditionally immortalizing primary podocytes, it is possible to overcome this limitation by the insertion of a thermosensitive switch, resulting in a cell culture model where cells proliferate at 33 °C and differentiate at 37 °C13,14. Although these ciPodocytes have high potential for podocyte research, there are limitations, like the lack of marker expression, dedifferentiated morphology, and failure to form foot processes15,16.
The differentiation of podocytes from patient-derived somatic cells enables the generation and comparison of diseased podocytes with healthy control cells ex vivo. This enables us to study podocyte damage due to mutations in podocyte-specific genes. Furthermore, working with hiPSCs has the potential for creating three-dimensional cell culture disease models, or rather organoids43,44. Co-culture of hiPSC-derived podocytes with other glomerular cells, like glomerular endothelial cells or mesangial cells, might lead to new insights regarding intercell communication in health and glomerular disease.
Moreover, the characterization and treatment of the patient-specific podocytes can be performed ex vivo in high-throughput analysis. The individualized approach opens the opportunity to investigate novel therapeutic targets for specific mutations and to perform individualized medicine in the future.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was funded by the Interdisciplinary Center for Clinical Research (IZKF) of Friedrich-Alexander University Erlangen-Nürnberg with the grant number M4-IZKF-F009 given to Janina Müller-Deile, and by Bundesministerium für Bildung und Forschung (BMBF) under the project name STOP-FSGS-Speed Translation-Oriented Progress to Treat FSGS, grant number 01GM2202D given to Janina Müller-Deile. We thank Annalena Kraus for the support in taking SEM images.
Materials
| Name | Company | Catalog Number | Comments |
| 0.2 µm sterile filter | Rotilab | P668.1 | for sterilization of differentiation medium |
| all-trans retinoic acid | Stem Cell Technologies | 72262 | supplement for differentiation |
| B27 supplement (50 x), serum free | Gibco | 17504044 | supplement for serum-free differentiation medium |
| BG iMatrix-511 Silk | biogems | RL511S | additional option of extracellular matrix reagent used in coating solution to coat cell culture plastics suitable for hiPSC culture |
| Bovine serum albumin (BSA) | Roth | 8076.4 | |
| CELLSTAR Filter Cap Cell Culture Flasks, T75, 250 mL | Greiner bio-one | 82050-856 | cell culture plastics suitable for fibroblast culture |
| CHIR99021 (5 mg) | Sigma-Aldrich | 252917-06-9 | supplement for differentiation |
| Corning Matrigel hESC qualified matrix | Corning | 354277 | additional option of extracellular matrix reagent used in coating solution to coat cell culture plastics suitable for hiPSC culture (solubilized basement membrane matrix) |
| countess Cell Counting Chamber Slides | Invitrogen | C10283 | to count cells |
| countess II FL Automated Cell Counter | Invitrogen | to count cells | |
| cryoPure tubes, 2 ml, QuickSeal screw cap, white | Sarstedt | 72380 | cryovials for freezing of cells |
| dimethyl sulfoxide (DMSO) | Roth | A994.1 | for fibroblast freezing medium |
| DMEM/F12 (1:1) (1 x) | Gibco | 11320074 | basic medium for differentiation |
| DMEM/F12 + Glutamax | Gibco | 10565018 | basic medium for fibroblast medium |
| EVOS M5000 Imaging System | Thermo Fisher Scientific | AMF5000 | phase contrast microscope |
| fetal bovine serum premium, inactivated (FCS) | PAN Biotech | P301902 | serum for fibroblast medium, fibroblast freezing medium and podocyte maintenance medium |
| fisherbrand Electroporation Cuvettes Plus, 4 mm gap, 800 µL capacity, sterile | Fisherbrand | FB104 | cuvette used for electroporation/episomal reprogramming of fibroblasts (4mm gap) |
| fluoromount-G Mounting Medium, with DAPI | Invitrogen | 00-4959-52 | mounting medium containing dapi |
| gauge needle (0.6 x 30 mm) | BD Microlance3 | 300700 | for separation of hiPSC colonies into small pieces |
| human Recombinant Activin A Protein | 78001.1 | Stem cell technologies | supplement for differentiation |
| human recombinant bone morphogenetic protein 7 (BMP7) | Peprotech | 120-03P | supplement for differentiation |
| human VEGF-165 Recombinant Protein | Thermo Scientific | PHC9394 | supplement for differentiation |
| insulin-transferrin-selenium (ITS -G) (100 x) | Gibco | 41400045 | supplement for podocyte maintenance medium |
| LB medium | Roth | X964.1 | for sterility test of hiPSC culture |
| lookOut Mycoplasma PCR Detection Kit | Sigma Aldrich | MP0035-1KT | commercial mycoplasma detection kit |
| microscope slides | Diagonal GmbH & Co.KG | 21,102 | |
| microtube 1.5 mL | Sarstedt | 72706400 | |
| mTeSR1 Complete Kit | Stem Cell Technologies | 85850 | basic medium for serum-free hiPSC culture medium |
| nalgene freezing container Mr.Frosty | Roth | AC96.1 | to ensure optimal freezing conditions |
| normal goat serum | abcam | ab 7481 | for preincubation solution and antibody diluent |
| nunc 24 well plates | Thermo Scientific | 142485 | cell culture plastics suitable for hiPSC culture |
| nunc 48 well plates | Thermo Scientific | 152640 | cell culture plastics suitable for hiPSC culture |
| nunc 6 well plates | Thermo Scientific | 140685 | cell culture plastics suitable for hiPSC culture |
| nunc EasYDish Dishes 100 mm | Thermo Scientific | 150466 | cell culture plastics suitable for hiPSC culture |
| nunc MicroWell 96-Well, Nunclon Delta-Treated, Flat-Bottom Microplate | Thermo Scientific | 167008 | cell culture plastics suitable for hiPSC culture |
| nutriFreez D10 Cryopreservation Medium | Sartorius | 05-713-1E | serum-free cryopreservation medium for cryopreservation of hiPSC and nephron progenitor cells |
| Opti-MEM | Gibco | 11058021 | electroporation medium |
| pCXLE-hMLN | Addgene | #27079 | plasmid for episomal reprogramming |
| pCXLE-hOCT3/4 plasmid | Addgene | #27077 | plasmid for episomal reprogramming |
| pCXLE-hSK plasmid | Addgene | #27078 | plasmid for episomal reprogramming |
| penicillin-streptomycin | Sigma-Aldrich | P4333-100ML | to avoid bacterial contamination |
| plastic coverslips | Sarstedt | 83.1840.002 | for immunofluorescent stainings of hiPSCs and hiPSC-derived podocytes |
| ROTI Histofix | Roth | P087.3 | commercial paraformaldehyde (4 %) for fixation of cells |
| RPMI 1640 + L-Glutamine | Gibco | 21875034 | basic medium for podocyte maintenance medium |
| staining chamber StainTray Black lid | Roth | HA51.1 | |
| stemPro Accutase Cell Dissociation Reagent | Gibco | A1110501 | enzymatic cell detachment solution used for dissociation of hiPSCs |
| sterile phosphate buffered saline (PBS) (1 x) | Gibco | 14190094 | used for washing and coating |
| sterile water | Roth | T1432 | |
| syringe without needle 20 mL | BD Plastipak | 300629 | to filter sterilize differentiation medium |
| TC dish 100 mm | Sarstedt | 8,33,902 | sterile cell culture plastics used for cutting the skin biopsy and fibroblast culture |
| TC dish 35 mm | Sarstedt | 8,33,900 | sterile cell culture plastics used for outgrowing fibroblasts from skin biopsy |
| triton X 100 | Roth | 3051.3 | for preincubation solution |
| trypan Blue Stain (0.4 %) for use with the Countess Automated Cell Counter | Invitrogen | T10282 | to count cells |
| trypsin-EDTA (10 x) | Biowest | X0930-100 | dissociation reagent used for fibroblasts and nephron progenitor cells |
| tube 15 mL | Greiner bio-one | 188271-N | |
| tube 50 mL | Greiner bio-one | 227261 | |
| vitronectin ACF | Sartorius | 05-754-0002 | extracellular matrix reagent used in coating solution to coat cell culture plastics suitable for hiPSC culture |
| Y-27632 dihydrochloride (10 mg) | Tocris | 1254 | to avoid apoptosis of hiPSCs during splitting |
| Primary antibodies | |||
| OCT4 | Stem Cell Technologies | 60093.1 | pluripotency marker, dilution 1:200 |
| SSEA-4 | Stem Cell Technologies | 60062FI.1 | pluripotency marker, dilution 1:100 |
| Ki67 | Abcam | ab15580 | proliferation marker, dilution 1:300 |
| synaptopodin | Proteintech | 21064-1-AP | podocyte-specific marker, dilution 1:200 |
| nephrin | Progen | GP-N2 | podocyte-specific marker, dilution 1:25 |
| podocin | proteintech | 20384-1-AP | podocyte-specific marker, dilution 1:100 |
| Secondary antibodies | |||
| goat anti-Guinea Pig IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Invitrogen | A21435 | secondary anditbody, dilution 1:1000 |
| alexa Fluor 647 Goat Anti-Rabbit SFX Kit, highly cross-adsorbed | Invitrogen | A31634 | secondary anditbody, dilution 1:1000 |
| donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | A21206 | secondary anditbody, dilution 1:1000 |
| goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Invitrogen | A21422 | secondary anditbody, dilution 1:1000 |
| goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | A11001 | secondary anditbody, dilution 1:1000 |
References
- Mundel, P., et al. Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Experimental Cell Research. 236 (1), 248-258 (1997).
- Grgic, I., et al. Imaging of podocyte foot processes by fluorescence microscopy. Journal of the American Society of Nephrology. 23 (5), 785-791 (2012).
- Grahammer, F., Schell, C., Huber, T. B. The podocyte slit diaphragm-from a thin grey line to a complex signalling hub. Nature Reviews Nephrology. 9 (10), 587-598 (2013).
- Deen, W. M., Lazzara, M. J., Myers, B. D. Structural determinants of glomerular permeability. American Journal of Physiology. Renal Physiology. 281 (4), F579-F596 (2001).
- Schell, C., Huber, T. B. The evolving complexity of the podocyte cytoskeleton. Journal of the American Society of Nephrology. 28 (11), 3166-3174 (2017).
- Muller-Deile, J., Schiffer, M. Podocyte directed therapy of nephrotic syndrome-can we bring the inside out. Pediatric Nephrology. 31 (3), 393-405 (2016).
- Boehlke, C., et al. Hantavirus infection with severe proteinuria and podocyte foot-process effacement. American Journal of Kidney Diseases. 64 (3), 452-456 (2014).
- Schiffer, M., et al. Pharmacological targeting of actin-dependent dynamin oligomerization ameliorates chronic kidney disease in diverse animal models. Nature Medicine. 21 (6), 601-609 (2015).
- Kopp, J. B., et al. Podocytopathies. Nature Reviews. Disease Primers. 6 (1), 68 (2020).
- Wiggins, R. C. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney International. 71 (12), 1205-1214 (2007).
- Mundel, P., Reiser, J., Kriz, W. Induction of differentiation in cultured rat and human podocytes. Journal of the American Society of Nephrology. 8 (5), 697-705 (1997).
- Jat, P. S., et al. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proceedings of the National Academy of Sciences. 88 (12), 5096-5100 (1991).
- Saleem, M. A., et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. Journal of the American Society of Nephrology. 13 (3), 630-638 (2002).
- Eto, N., et al. Podocyte protection by darbepoetin: preservation of the cytoskeleton and nephrin expression. Kidney International. 72 (4), 455-463 (2007).
- Krtil, J., Platenik, J., Kazderova, M., Tesar, V., Zima, T. Culture methods of glomerular podocytes. Kidney & Blood Pressure Research. 30 (3), 162-174 (2007).
- Chittiprol, S., Chen, P., Petrovic-Djergovic, D., Eichler, T., Ransom, R. F. Marker expression, behaviors, and responses vary in different lines of conditionally immortalized cultured podocytes. American Journal of Physiology. Renal Physiology. 301 (3), F660-F671 (2011).
- Shih, N. Y., et al. CD2AP localizes to the slit diaphragm and binds to nephrin via a novel C-terminal domain. The American Journal of Pathology. 159 (6), 2303-2308 (2001).
- Yan, K., Khoshnoodi, J., Ruotsalainen, V., Tryggvason, K. N-linked glycosylation is critical for the plasma membrane localization of nephrin. Journal of the American Society of Nephrology. 13 (5), 1385-1389 (2002).
- Sir Elkhatim, R., Li, J. Y., Yong, T. Y., Gleadle, J. M. Dipping your feet in the water: podocytes in urine. Expert Review of Molecular Diagnostics. 14 (4), 423-437 (2014).
- Camici, M. Urinary detection of podocyte injury. Biomedicine & Pharmacotherapy. 61 (5), 245-249 (2007).
- Muller-Deile, J., et al. Overexpression of preeclampsia induced microRNA-26a-5p leads to proteinuria in zebrafish. Scientific Reports. 8 (1), 3621 (2018).
- Schenk, H., et al. Removal of focal segmental glomerulosclerosis (FSGS) factor suPAR using CytoSorb. Journal of Clinical Apheresis. 32 (6), 444-452 (2017).
- Petermann, A., Floege, J. Podocyte damage resulting in podocyturia: a potential diagnostic marker to assess glomerular disease activity. Nephron. Clinical Practice. 106 (2), c61-c66 (2007).
- Vogelmann, S. U., Nelson, W. J., Myers, B. D., Lemley, K. V. Urinary excretion of viable podocytes in health and renal disease. American Journal of Physiology. Renal Physiology. 285 (1), F40-F48 (2003).
- Petermann, A. T., et al. Podocytes that detach in experimental membranous nephropathy are viable. Kidney International. 64 (4), 1222-1231 (2003).
- Sakairi, T., et al. Conditionally immortalized human podocyte cell lines established from urine. American Journal of Physiology. Renal Physiology. 298 (3), F557-F567 (2010).
- Rauch, C., et al. Differentiation of human iPSCs into functional podocytes. PLoS One. 13 (9), e0203869 (2018).
- Musah, S., et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nature Biomedical Engineering. 1, 0069 (2017).
- Takahashi, K., Okita, K., Nakagawa, M., Yamanaka, S. Induction of pluripotent stem cells from fibroblast cultures. Nature Protocols. 2 (12), 3081-3089 (2007).
- Takahashi, K., Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126 (4), 663-676 (2006).
- Teshigawara, R., Cho, J., Kameda, M., Tada, T. Mechanism of human somatic reprogramming to iPS cell. Laboratory Investigation. 97 (10), 1152-1157 (2017).
- Bang, J. S., et al. Optimization of episomal reprogramming for generation of human induced pluripotent stem cells from fibroblasts. Animal Cells and Systems. 22 (2), 132-139 (2018).
- Okita, K., et al. A more efficient method to generate integration-free human iPS cells. Nature Methods. 8 (5), 409-412 (2011).
- Musah, S., Dimitrakakis, N., Camacho, D. M., Church, G. M., Ingber, D. E. Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a Glomerulus Chip. Nature Protocols. 13 (7), 1662-1685 (2018).
- Burt, M., Bhattachaya, R., Okafor, A. E., Musah, S. Guided differentiation of mature kidney podocytes from human induced pluripotent stem cells under chemically defined conditions. Journal of Visualized Experiments. (161), e61299 (2020).
- Vangipuram, M., Ting, D., Kim, S., Diaz, R., Schule, B. Skin punch biopsy explant culture for derivation of primary human fibroblasts. Journal of Visualized Experiments. (77), e3779 (2013).
- Hoffding, M. K., Hyttel, P. Ultrastructural visualization of the Mesenchymal-to-Epithelial Transition during reprogramming of human fibroblasts to induced pluripotent stem cells. Stem Cell Research. 14 (1), 39-53 (2015).
- Bharathan, S. P., et al. Systematic evaluation of markers used for the identification of human induced pluripotent stem cells. Biology Open. 6 (1), 100-108 (2017).
- Scholzen, T., Gerdes, J. The Ki-67 protein: from the known and the unknown. Journal of Cellular Physiology. 182 (3), 311-322 (2000).
- Sun, X., Kaufman, P. D. Ki-67: more than a proliferation marker. Chromosoma. 127 (2), 175-186 (2018).
- Vaz, I. M., et al. Chromosomal aberrations after induced pluripotent stem cells reprogramming. Genetics and Molecular Biology. 44 (3), 20200147 (2021).
- Reiser, J., Altintas, M. M. Podocytes. F1000Research. 5, 114 (2016).
- Ohmori, T., et al. Impaired NEPHRIN localization in kidney organoids derived from nephrotic patient iPS cells. Scientific Reports. 11 (1), 3982 (2021).
- Morizane, R., Bonventre, J. V. Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nature Protocols. 12 (1), 195-207 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved