Method Article
Hybrid Ensemble and Single-molecule Assay to Image the Motion of Fully Reconstituted CMG
In This Article
Summary
We report a hybrid ensemble and single-molecule assay to directly image and quantify the motion of fluorescently labeled, fully reconstituted Cdc45/Mcm2-7/GINS (CMG) helicase on linear DNA molecules held in place in an optical trap.
Abstract
Eukaryotes have one replicative helicase known as CMG, which centrally organizes and drives the replisome, and leads the way at the front of replication forks. Obtaining a deep mechanistic understanding of the dynamics of CMG is critical to elucidating how cells achieve the enormous task of efficiently and accurately replicating their entire genome once per cell cycle. Single-molecule techniques are uniquely suited to quantify the dynamics of CMG due to their unparalleled temporal and spatial resolution. Nevertheless, single-molecule studies of CMG motion have thus far relied on pre-formed CMG purified from cells as a complex, which precludes the study of the steps leading up to its activation. Here, we describe a hybrid ensemble and single-molecule assay that allowed imaging at the single-molecule level of the motion of fluorescently labeled CMG after fully reconstituting its assembly and activation from 36 different purified S. cerevisiae polypeptides. This assay relies on the double functionalization of the ends of a linear DNA substrate with two orthogonal attachment moieties, and can be adapted to study similarly complex DNA-processing mechanisms at the single-molecule level.
Introduction
DNA replication in eukaryotes is carried out by a dynamic protein complex known as the replisome1. A key component of this complex is the eukaryotic replicative helicase Cdc45/Mcm2-7/GINS (CMG), which drives and centrally organizes the replisome, leading the way at the front of replication forks1,2. Obtaining a deep quantitative understanding of the dynamics of CMG is therefore critical to understanding the dynamics of the replisome. Such an understanding could be acquired with single-molecule techniques, which are uniquely suited to study molecular motors, such as CMG, due to their unmatched spatial and temporal resolution, and can provide us with an unparalleled quantitative understanding of their function, stochasticity, and dynamics2,3,4,5,6,7,8,9.
In vivo, CMG is loaded and activated in temporally separated fashion to ensure that replication occurs only once per cell cycle1,10,11. First, in the G1 phase of the cell cycle, a set of proteins known as loading factors loads the first component of CMG, the Mcm2-7 hexameric complex, onto dsDNA12,13,14,15,16 in the form of double hexamers with a head-to-head configuration15,17,18. In the specific case of yeast, this initial process occurs at specific DNA sequences known as origins of replication1. Although Mcm2-7 is the motor core of the replicative helicase, it is by itself unable to unwind DNA19 without the two helicase-activating factors Cdc45 and GINS, which need to be recruited to the loaded Mcm2-7 to give rise to fully active CMG11,19,20,21. The process of helicase activation takes place in the S phase of the cell cycle and starts with the selective phosphorylation of Mcm2-7 double hexamers by the cell cycle-regulated kinase DDK22,23,24. These phosphorylation events facilitate the recruitment of Cdc45 and GINS to the Mcm2-7 double hexamers10,22,23,24,25,26 by a second set of proteins known as firing factors10,11,26. The binding of Cdc45 and GINS gives rise to two sister CMG helicases, which initially enclose both strands of the parental DNA and are still located in a head-to-head configuration11,27. In the final activation step, the firing factor Mcm10 catalyzes the ATP hydrolysis-dependent extrusion of one DNA strand from each sister CMG11. After strand extrusion, sister CMG helicases bypass and separate from each other by translocating along ssDNA in an ATP hydrolysis-dependent manner11,20,21,28, unwinding DNA by sterically excluding the non-translocation strand29. This entire process has been fully reconstituted in vitro from a minimal set of 36 purified S. cerevisiae polypeptides10,11.
Despite the exquisite in vivo regulation of CMG assembly and activation described above, in vitro reconstituted single-molecule motion studies of CMG2,30,31,32,33,34 have thus far relied on pre-activated CMG purified as a complex from cells20,21, missing all the steps prior to its activation and the bidirectional nature of its motion. This pre-activated CMG approach has been the gold standard in the single-molecule field partly due to the biochemical complexity of the fully reconstituted CMG assembly reaction10,11. This biochemical reaction has been challenging to translate from the bulk biochemical level to the single-molecule level for several reasons. First, to maximize reaction efficiencies, the loading and firing factors needed for CMG assembly and activation are required at concentrations in the range of 10-200 nM10,11,27. These ranges of concentration correspond to the high end of what most single-molecule techniques can tolerate, especially when using fluorescently labeled components35. Finally, CMG has evolved to cruise through thousands of base pairs in a cell36,37,38,39. Therefore, to study its motion at a biologically relevant spatial scale, one requires long DNA substrates (typically of lengths in the order of tens of kilobases)30,31,34,40,41,42. Employing such long DNA substrates poses the additional challenge that the longer the DNA substrate is, the more potential non-specific binding sites for proteins and protein aggregates it has. In the case of CMG, the latter point is particularly important, as several of the loading and firing factors involved in CMG assembly and activation contain intrinsically disordered regions43 and are aggregation-prone.
Here, we report a hybrid ensemble and single-molecule assay that allowed the observation and quantification of the motion of CMG after fully reconstituting its assembly and activation from 36 purified S. cerevisiae polypeptides28. This assay relies on the double functionalization of both ends of a DNA substrate with two orthogonal attachment moieties: desthiobiotin and digoxigenin2 (Figure 1A). The first moiety, desthiobiotin, is used to reversibly bind the DNA substrate to streptavidin-coated magnetic beads44 (Figure 1B). Following this, fluorescently labeled CMG is assembled and activated onto the bead-bound DNA, and a magnetic rack is used to purify and wash the resulting magnetic bead-bound DNA:CMG complexes (Figure 1C). In doing so, the excess protein that would otherwise aggregate on the DNA substrate is removed; this provides virtually aggregation-free DNA:CMG complexes. Intact complexes are then eluted from the magnetic beads by the addition of a molar excess of free biotin, which can outcompete the desthiobiotin-streptavidin interaction (Figure 1D). Individual DNA:CMG complexes are then bound between two optically trapped polystyrene beads coated with anti-digoxigenin antibody (anti-Dig); for this step, the second moiety on the DNA, digoxigenin, is used as it can bind to anti-Dig even in a buffer solution containing an excess of free biotin (Figure 1E). Once the DNA:CMG complex is held in place in the optical trap, the motion of fluorescently labeled CMG is imaged with a confocal scanning laser (Figure 1F). We anticipate that this assay can be easily adapted for the study at the single-molecule level of similarly complex DNA:protein interactions.
Protocol
1. Synthesis of doubly functionalized linear DNA substrate and binding to magnetic beads
- Dual functionalization of DNA substrate with desthiobiotin and digoxigenin moieties
- Linearize 20 µg of 23.6 kb plasmid pGL50-ARS1 (containing a natural ARS1 origin of replication, cloned in house and available upon request) with 200 units of restriction enzyme AflII for 16 h at 37 °C in a final volume of 200 µL of 1x buffer (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 100 µg/ml BSA, pH 7.9).
NOTE: This step can be reduced to 4 h without reducing the yield, if more convenient. - Inactivate AflII by incubating the reaction at 65 °C for 20 min. Blunt the resulting 4-nucleotide TTAA overhangs by supplementing the 200 µL linearization reaction with 60 units of Klenow Fragment (3' to 5' exo-) polymerase, 17 µL of 10x buffer (500 mM NaCl, 100 mM Tris-HCl, 100 mM MgCl2, 10 mM DTT, pH 7.9), 33 µM D-Desthiobiotin-7-dATP, 33 µM Digoxigenin-11-dUTP, 33 µM dCTP, 33 µM dGTP, and ultrapure water to a final volume of 370 µM. Incubate the reaction at 37 °C for 30 min.
NOTE: In this step, the Klenow Fragment blunts the overhangs at both ends of the linearized DNA by incorporating two D-Desthiobiotin-7-dATP and two Digoxigenin-11-dUTP nucleotides at each end. - Supplement the reaction with 10 mM EDTA and inactivate the Klenow Fragment by incubating the reaction at 75 °C for 20 min. Bring the reaction volume to 400 µL with ultrapure water.
- Take four S-400 spin columns, vortex them for at least 30 s to resuspend the resin, and then centrifuge them for 1 min at 735 x g to remove the storage buffer. Transfer the columns to clean 1.5 mL tubes.
- Immediately, add 100 µL of the DNA solution to each column and centrifuge them for 2 min at 735 x g. The DNA is now in the flow-through, and the columns may be discarded.
- Pool together the flow-through of the four columns, measure the volume with a pipette and measure the DNA concentration with a spectrophotometer. This will be used to calculate the amount of DNA bound to the beads.
- Linearize 20 µg of 23.6 kb plasmid pGL50-ARS1 (containing a natural ARS1 origin of replication, cloned in house and available upon request) with 200 units of restriction enzyme AflII for 16 h at 37 °C in a final volume of 200 µL of 1x buffer (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 100 µg/ml BSA, pH 7.9).
- Binding doubly functionalized linear DNA to streptavidin-coated magnetic beads
- Vortex M-280 streptavidin-coated magnetic beads for 30 s to resuspend them.
- Transfer 4 mg of resuspended M-280 streptavidin-coated magnetic beads to a clean 1.5 mL tube. Place the tube in a magnetic rack, wait 1 min for the beads to be collected, and remove the storage buffer.
- Add 1 mL of 1x buffer A (5 mM Tris-HCl pH 7.5, 0.5 mM EDTA, and 1 M NaCl) to the beads and resuspend by vortexing for 5 s. Incubate the beads at room temperature for 5 min.
NOTE: All buffers should be sterile filtered. Make a large volume of 2x buffer A, 1x buffer B (see below) and 1x buffer C (see below) beforehand to save time (recommended). These buffers are the ones recommended by the manufacturer of the magnetic beads and can be stored at - 20°C. - Place the tube in a magnetic holder, wait 1 min for the beads to be collected, and remove buffer A.
- Resuspend the beads in 400 µL of 2x buffer A by pipetting, and then add 400 µL of functionalized DNA solution (note that this dilutes the 2x buffer A to a final concentration of 1x, which is optimal for DNA binding to the beads). Mix gently by pipetting.
- Incubate the bead/DNA mixture overnight at 4 °C with end-over-end rotation to allow the functionalized DNA to bind to the beads.
- Use the magnetic rack to remove the supernatant (do not discard before measuring its volume with a pipette and the concentration of unbound DNA with a spectrophotometer) and wash the beads 2x with 500 µL of buffer B (10 mM HEPES-KOH pH 7.6, 1 mM EDTA, and 1 M KOAc) to remove non-specifically bound DNA. Wash the beads 2x with 500 µL of buffer C (10 mM HEPES-KOH pH 7.6 and 1 mM EDTA), which is the storage buffer recommended by the manufacturer.
- Calculate the total amount of DNA bound to the magnetic beads by comparing the total amount of DNA added to the beads with the amount of DNA left in the supernatant. The yield should be in the range of 2.3-2.9 mg of DNA (~150-190 fmol) per mg of magnetic beads. If lower yields are obtained, check that the S400 columns are not dry before using them. Constantly monitor the DNA integrity by gel electrophoresis to ensure that the initial plasmid DNA substrate is not degraded.
- Resuspend the beads in 300 µL of buffer C and store at 4 °C. To prevent nicking, make 4 single-use aliquots of 1 mg of magnetic beads, and do not store the DNA for longer than 2 weeks.
2. Hybrid ensemble and single-molecule assay to image and quantify the motion of fully reconstituted CMG with correlative dual-beam optical tweezers and confocal microscopy
- Ensemble assembly and activation of fluorescently labeled CMG onto magnetic bead-bound DNA (Figure 1C).
NOTE: CMG assembly and activation reactions were carried out in two stages: Mcm2-7 loading and phosphorylation, and CMG assembly and activation. Unless otherwise specified, all buffer exchange steps were conducted with the help of a magnetic rack by allowing the beads to be collected by the magnet for 1 min and then removing the supernatant. All incubations were conducted in a temperature-controlled heat block with a lid to prevent photobleaching of fluorescent proteins. If a lid is not available, the tubes should be covered in tin foil. The purification and fluorescent labeling of all the proteins employed in this protocol have been carried out as previously described10,11,28.- Mcm2-7 loading and phosphorylation
- Take 1 mg of DNA-bound streptavidin-coated magnetic beads and remove the storage buffer (buffer C).
- Wash the beads with 200 µL of loading buffer (25 mM HEPES-KOH pH 7.6, 100 mM K glutamate, 10 mM MgOAc, 0.02% NP40 substitute, 10% glycerol, 2 mM DTT, 100 µg/mL BSA, and 5 mM ATP).
- Remove the loading buffer and resuspend the beads in 75 µL of loading buffer. Mix gently by pipetting.
- Add ORC at a final concentration of 37.5 nM to the bead-bound DNA and incubate the reaction for 5 min at 30 °C with 800 rpm agitation.
- Add Cdc6 at a final concentration of 50 nM and incubate the reaction for 5 min at 30 °C with 800 rpm agitation.
- Add Mcm2-7/Cdt1 (or fluorescently labeled Mcm2-7JF646-Halo-Mcm3/Cdt1) at a final concentration of 100 nM and incubate the reaction for 20 min at 30 °C with 800 rpm agitation.
- Add DDK at a final concentration of 100 nM and incubate the reaction for 30 min at 30 °C with 800 rpm agitation.
- Remove the supernatant and wash the bead-bound DNA (which now contains phosphorylated Mcm2-7 hexamers) with 200 µL of high-salt wash (HSW) buffer (25 mM HEPES-KOH pH 7.6, 300 mM KCl, 10 mM MgOAc, 0.02% NP40 substitute, 10% glycerol, 1 mM DTT, and 400 µg/mL BSA). Mix by pipetting, ensuring the beads are fully resuspended in the buffer, and no clumps are visible.
- Remove the HSW buffer and wash the beads once with 200 µL of CMG buffer (25 mM HEPES-KOH pH 7.6, 250 mM K glutamate, 10 mM MgOAc, 0.02% NP40 substitute, 10% glycerol, 1 mM DTT, and 400 µg/mL BSA).
- Assembly and activation of fluorescently labeled CMG
- Remove the CMG buffer and resuspend the DNA-bound beads in 50 µL of CMG buffer supplemented with 5 mM ATP.
- Add 50 nM Dpb11, 200 nM GINS, 30 nM Polɛ, 20 nM S-CDK, 50 nM Cdc45LD555, 30 nM Sld3/7, 55 nM Sld2, and 10 nM Mcm10 to the bead-bound DNA. For this step, mix all the proteins in one tube immediately before adding them to the DNA and place it on ice. Add the resuspended bead-bound DNA to the protein mix. Incubate the reaction for 15 min at 30 °C with 800 rpm agitation.
- Wash the beads 3x with 200 µL of HSW buffer. Wash the beads once with 200 µL of CMG buffer.
- Elution of intact DNA:CMG complexes from magnetic beads (Figure 1D)
- Remove CMG buffer and elute DNA:CMG complexes from the magnetic beads by resuspending the CMG-containing DNA-bound magnetic beads in 200 µL of elution buffer (CMG buffer supplemented with 10 mM biotin). Incubate at room temperature for 1 h with 800 rpm agitation.
- Place the tube in a magnetic rack and allow the beads to be collected for 5 min. Carefully collect the supernatant (which now contains the eluted DNA:CMG complexes) without disrupting the settled beads and transfer it to a new tube.
- To ensure that no beads remain in the solution (as magnetic beads left in solution can interfere with the optical trapping part of the assay), place the collected supernatant again in a magnetic rack and allow any remaining beads to be collected for another 5 min. Carefully collect the supernatant and transfer it to a new tube.
- Add 1400 µL of CMG buffer to the 200 µL of supernatant. The sample is now ready for single-molecule imaging.
- Mcm2-7 loading and phosphorylation
- Single-molecule imaging of fully reconstituted CMG with correlative dual-beam optical tweezers and confocal microscopy (Figure 1E-G).
NOTE: The single-molecule part of the assay is conducted in a commercial setup that combines dual-beam optical tweezers with confocal microscopy and microfluidics45,46 (Figure 1E-G), but may also be conducted in a home-built setup. The commercial setup used here is equipped with a microfluidic flow cell with five inlets (referred to as Channels 1-5, respectively) and one outlet (Figure 1E). In the steps below, all button and/or panel names are specific to the software that is provided with this commercial setup.- Use the five inlets in the commercial setup as follows: Inject Channels 1-3 from the left and use them for bead trapping (Channel 1), DNA-protein complex binding (Channel 2), and checking for the presence of CMG (Channel 3). Use Channels 4 and 5 as protein reservoirs and buffer exchange locations. Before each experiment, passivate the microfluidic flow cell for at least 30 min with 1 mg/mL bovine serum albumin, followed by 0.5% Pluronic F-127 dissolved in ultrapure water.
- Ensure that the content of each channel in each experiment is as described (Figure 1E):
Channel 1: Anti-digoxigenin-coated polystyrene beads (2.06 µm diameter) diluted 1:50 in PBS;
Channel 2: DNA:CMG complexes eluted from magnetic beads;
Channel 3: Imaging Buffer (CMG buffer supplemented with 2 mM 1,3,5,7 cyclooctatetraene, 2 mM 4-nitrobenzylalchohol, and 2 mM Trolox);
Channels 4 and 5: Imaging Buffer supplemented with 25 nM RPA, 10 nM Mcm10, and either 5 mM ATP, 5 mM ATPγS, or no nucleotide.
- Ensure that the content of each channel in each experiment is as described (Figure 1E):
- Before the experiment, adjust the trapping laser power to achieve a stiffness of 0.3 pN/nm in both traps33,46 by clicking on the Re-calibrate button in the Power Controls panel of the software. Set the confocal pixel size to 50 x 50 nm, the image size to 160 x 18 pixels, the illumination time per pixel to 0.2 ms, and the frame rate to 5 s in the Image Scan panel of the software. Set the temperature control to 30 °C in the Temperature panel.
NOTE: In addition, we recommend setting the maximum stage velocity (used to move between channels) to 0.2 mm/s to minimize CMG dissociation from the DNA by the drag force. - Flow all solutions into the flow cell at a constant pressure of 0.5 bar in the injection port (which can be set up in the Pressure Bar panel of the software). Then, turn off the flow in channels 4 and 5. After initially flowing all solutions, reduce the pressure to 0.2 bar.
- Move trapping lasers to Channel 1 until one bead is caught in each optical trap. Move trapped beads to Channel 2 by moving the joystick while pressing on the trigger. Then, fish a DNA:CMG complex by moving the right bead towards and away from the left bead using the joystick without pressing the trigger, until a DNA tether is trapped (which is known by monitoring the force-extension curve of the trapped DNA47 in the FD Viewer panel of the software).
- For DNA tethering, ensure to input the DNA length and the correct temperature value in the Reference Model panel, which will automatically plot a theoretical extensible worm-like chain model of the DNA construct in the FD Viewer panel. If a single DNA molecule is caught between the two beads, the force-extension behavior of the DNA should closely match the plotted extensible worm-like chain model. Ensure that this is the case by monitoring the experimental force-extension curve, which the software will automatically plot on top of the theoretical one.
NOTE: Deviations from the theoretical model can indicate the presence of multiple DNA molecules bound between the beads and/or the presence of protein aggregates bound to the DNA and compacting it. To prevent the force-mediated dissociation of proteins from the DNA, do not exceed a tension of 10 pN during this initial test.
- For DNA tethering, ensure to input the DNA length and the correct temperature value in the Reference Model panel, which will automatically plot a theoretical extensible worm-like chain model of the DNA construct in the FD Viewer panel. If a single DNA molecule is caught between the two beads, the force-extension behavior of the DNA should closely match the plotted extensible worm-like chain model. Ensure that this is the case by monitoring the experimental force-extension curve, which the software will automatically plot on top of the theoretical one.
- Move the beads to Channel 3 and immediately stop the flow in all channels. Take a one-frame test scan in Channel 3 to confirm the presence of CMG, illuminating with a 561 nm laser at a power of 4 µW as measured at the objective. Adjust imaging laser powers in the Excitation lasers panel. If present, CMG will appear as two-dimensional diffraction-limited spots as shown in Figure 1G and Figure 2A.
- If no DNA can be caught, check channel 2 for bubbles, which can slow down the flow in channel 2 (compared to channels 1 and 3). If bubbles are present, increase the pressure in channel 2 to 1 bar to try to flush the bubbles through.
- If CMG is present, move the DNA tether to either channel 4 or 5 for imaging; otherwise, turn the flow back on, discard the beads, and go back to step 2.2.4.
- In channels 4 or 5, adjust the distance between the beads to achieve an initial tension of 2 pN in the DNA tether. For this, input 2 pN in the Force Spectroscopy panel and click on the Enable button to start a force clamp. Once the initial tension reaches 2 pN, disable the force clamp before imaging by clicking on the Enable button in the Force Spectroscopy panel.
- Image CMGCdc45-LD555 every 5 s with a 561 nm laser at a power of 4 µW as measured at the objective (Figure 1F). For this, click the Start Scan button in the Image scan panel. In such scans, CMG will show as two-dimensional diffraction-limited spots, such as the example in Figure 1G.
- If CMG is observed but it does not seem to move before bleaching, test all the proteins used for nuclease activity, as nicks on the DNA will cause CMG to dissociate from DNA41, severely reducing its apparent processivity.
NOTE: A laser power of 4 µW should give consistent results if using the commercial microscope used here. If using a home-build setup, we recommend estimating the optimal laser power empirically. An optimal laser power should give enough signal from the fluorophore to localize it with the desired precision, and a fluorophore lifetime close to the desired time range of the experiment.
NOTE: Although this publication focuses on the hybrid ensemble and single-molecule assay, we have previously published a comprehensive description of the analysis of the data generated with this assay48.
- If CMG is observed but it does not seem to move before bleaching, test all the proteins used for nuclease activity, as nicks on the DNA will cause CMG to dissociate from DNA41, severely reducing its apparent processivity.
- Use the five inlets in the commercial setup as follows: Inject Channels 1-3 from the left and use them for bead trapping (Channel 1), DNA-protein complex binding (Channel 2), and checking for the presence of CMG (Channel 3). Use Channels 4 and 5 as protein reservoirs and buffer exchange locations. Before each experiment, passivate the microfluidic flow cell for at least 30 min with 1 mg/mL bovine serum albumin, followed by 0.5% Pluronic F-127 dissolved in ultrapure water.
Results
When carried out correctly, the protocol described here should yield virtually aggregate-free DNA:CMG complexes. An aggregation-free reaction should not clog any of the channels in the microfluidic flow cell, and it should be possible to stretch the trapped DNA molecules to an end-to-end extension within 10% of its contour length without breaking the DNA. Conversely, if there is aggregation in the reaction, DNA molecules may sometimes become compacted by the aggregates, often causing DNA breakage if stretched. A good way to recognize protein aggregates is by looking at the 2D scans of the DNA. In an aggregation-free reaction, CMG appears as discrete, symmetrical diffraction-limited spots sparsely crowding the DNA, such as the ones in the scans shown in Figure 2A. On the contrary, aggregates are less discrete, sometimes asymmetrical blobs crowding a larger length of the DNA, like those in the scans shown in Figure 2B. Furthermore, if the assay is successfully executed, and high purity of the purified proteins is achieved, long-range motion of CMG in the presence of ATP will be seen, as observed in the kymograph shown in Figure 2C.
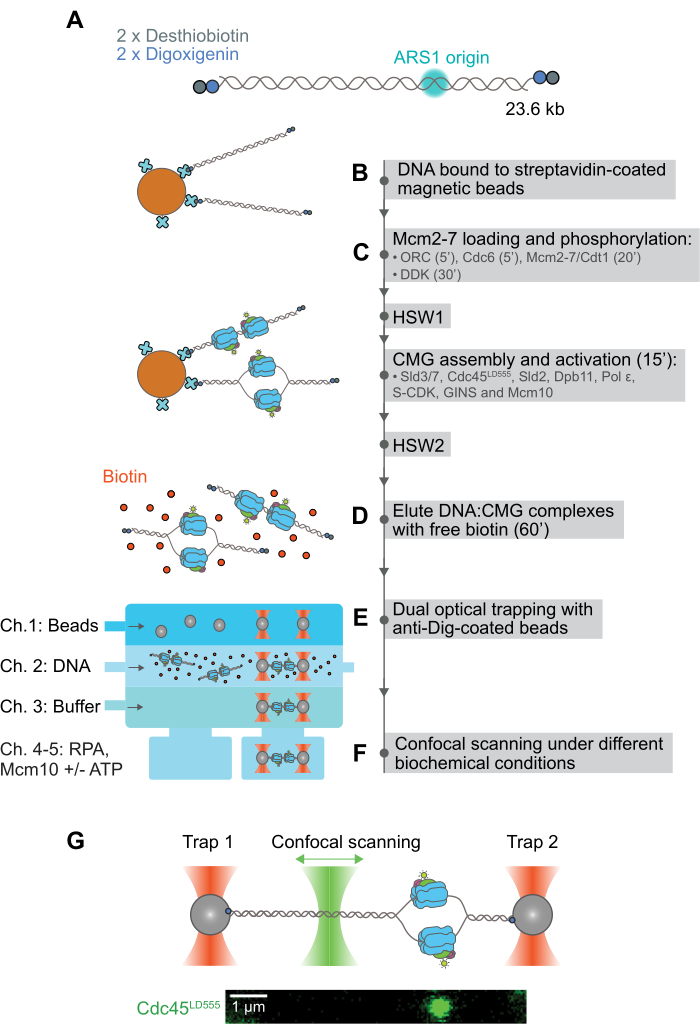
Figure 1: Pictorial description of hybrid ensemble and single-molecule assay to image and quantify the motion of fully reconstituted CMG. (A) A 23.6 kb linear DNA containing a naturally occurring ARS1 origin of replication is doubly functionalized at both ends with desthiobiotin and digoxigenin moieties. (B) Doubly functionalized DNA is bound to streptavidin-coated magnetic beads through its desthiobiotin moieties. (C) CMG is stepwise assembled and activated on the magnetic bead-bound DNA with different washing steps included to remove excess unbound protein and protein aggregates. (D) Intact DNA:CMG complexes are then eluted from the magnetic beads by the addition of an excess of free biotin, which outcompetes the desthiobiotin-streptavidin interaction. (E) Individual DNA:CMG complexes are bound between two anti-digoxigenin-coated optically trapped polystyrene beads with the help of a microfluidic flow cell. Note that the Dig-anti-Dig interaction is orthogonal to biotin-avidin interactions, so it is not affected by the presence of free biotin. (F) Once held in place by the optical tweezers, DNA:CMG complexes are transferred into different buffer conditions, where the DNA plane is then scanned with a confocal scanning laser to image the motion of CMG along the DNA over time. (G) The top panel shows a diagram of a DNA:CMG complex held in place between two optically trapped anti-Dig-coated polystyrene beads being scanned by a confocal scanning laser. The bottom panel shows an example 2D scan of CMG bound to a DNA held in place with an optical trap. The DNA is unlabeled in these experiments, but it can be thought of as a horizontal line running through the middle of the image. This figure has been modified from28. Please click here to view a larger version of this figure.
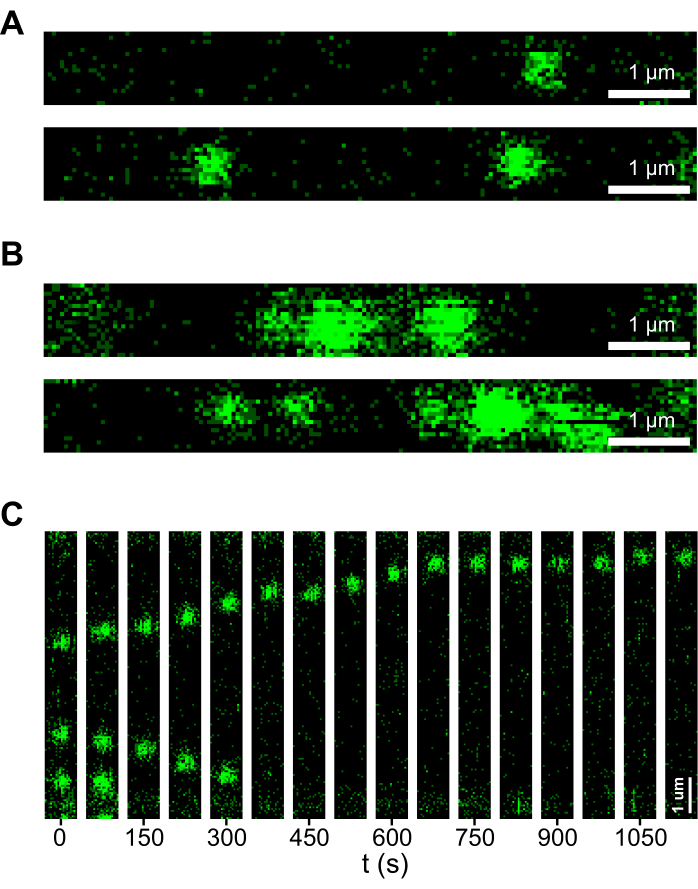
Figure 2: Examples of data from a successful and an unsuccessful experiment. (A) Example 2D scans of a CMG-containing DNA in an aggregation-free sample. In both scans, CMG shows symmetrical and discrete diffraction-limited spots sparsely distributed along the DNA. (B) Example 2D scans of a CMG-containing DNA in a sample containing aggregates. In both scans, CMG forms less symmetrical blobs crowding the DNA. (C) Kymograph showing the position on the DNA of CMG diffraction-limited spots over time in the presence of ATP, showing the long-range motion of CMG complexes. Please click here to view a larger version of this figure.
Discussion
Critical steps and important reagent quality checks
The critical steps and biological reagent quality checks in the assay are highlighted here. First, the purity of the proteins used is important because DNA degradation caused by even small nuclease contaminants in the protein samples will adversely affect the data. This is because only intact (or partially nicked) DNA molecules can be trapped in the dual-beam optical tweezers. More importantly, nicks on the DNA will cause CMG to dissociate41, complicating the observation of CMG's long-range motion. We strongly recommend testing each purified protein for nuclease activity, as well as constantly monitoring the integrity of the starting plasmid substrate to ensure that nicking is reduced to a minimum. The second important step is the careful removal of the magnetic beads following the elution of DNA:CMG complexes. Supernatant removal in this step should be conducted slowly so as not to perturb the collected beads. If magnetic beads are left in the sample flown into the optical tweezers, they will often hit the optically trapped polystyrene beads, causing them to escape the optical trap and complicating the data acquisition. Finally, DNA:CMG complexes should be handled carefully in the optical tweezers. To this end, we recommend not increasing the tension of the DNA above 10 pN, as the application of force may dissociate CMG from the DNA. Furthermore, moving between channels in the microfluidic flow cell should be done as slowly as possible (~ 0.2 mm/s) to prevent the resulting drag forces from dissociating CMG from the DNA.
Modifications of the method
There are several steps of the assay that could be modified. For instance, we have shown that the elution time can be reduced from 60 min to 30 min without significantly affecting the elution yield. In addition, we recommend supplementing the elution buffer with a low (below 1 mM) concentration of either ATP or ATPγS to prevent CMG from diffusing off the DNA ends as well as to generally stabilize CMG28. Further, although the buffer compositions and protein concentrations we report here are based on those employed in prior ensemble biochemical and single-molecule work11,18, the assay we describe is fully compatible with other protocols to assemble CMG26,27. Therefore, any biochemical advancement reported to increase the efficiency of CMG assembly or activation could and should be implemented in the bulk part of the assay to increase the yield. Finally, increasing the time between frames increases the total time in which CMG can be imaged, facilitating the observation of long-range CMG motion before fluorophore bleaching.
Limitations of the method
The hybrid method we describe is limited in that one can only image CMG following its activation in bulk. Further work will be required to observe the activation of CMG in real-time. Another important limitation is that, while we expect CMG to be assembled in pairs17,26,27, the total number of CMG complexes per DNA that we observe is mostly one28, suggesting that CMG, or at least Cdc45 is dissociating from the DNA during the handling of the sensitive DNA:CMG complexes. Reducing the number of handling steps prior to the single-molecule imaging, as well as developing better passivation strategies for the plastic tubing and glass of the microfluidic flow cell are poised to increase this yield.
Significance of the method
Single-molecule motion studies of CMG have thus far employed pre-activated CMG purified as a complex from cells. While relatively simpler, this pre-activated CMG approach is limited in that it misses any of the steps leading up to CMG activation, as well as the bidirectional nature of CMG and replisome motion. On the other hand, the full reconstitution of CMG assembly and activation has the potential to study any pre-activation steps, as well as to study CMG motion in a bidirectional manner. Nevertheless, this approach is harder to translate from the bulk biochemical level to the single-molecule level, as it involves a lot more purified protein factors and steps. The assay we describe here has helped to overcome these challenges by allowing us to image the motion of fully reconstituted CMG at the single-molecule level, allowing us to access some previously missed pre-activation dynamics28. Additionally, although we mostly see one CMG per DNA, we were able to observe several instances of two CMG complexes moving in opposite directions, and we could even capture the initial separation of sister CMGs from one another28, providing some insights into the establishment of bidirectional replication.
Another advantage of this assay compared to previous CMG motion lies in the fully double-stranded nature of the DNA substrate we employ (Figure 1A). In previous pre-activated CMG work, the most common way of binding CMG to the DNA substrate is through a 3' ssDNA flap. This results in a DNA construct that cannot be easily torsionally constrained and thus prohibits the study of the role of supercoiling in replisome progression. Conversely, the new approach we describe here could have the potential to be adapted to study the role of torque in this process, as the DNA substrate used is completely double stranded.
Broader applications of the method
The hybrid assay described will pave the way toward the full reconstitution of a complete eukaryotic replisome, allowing us and others to observe and quantify the important dynamics that allow the replisome to succeed at all its different tasks. DNA replication aside, the assay we report represents an important advancement in translating a complicated biochemical reaction from the bulk biochemical to the single-molecule level. We anticipate that this assay can be easily modified to study similarly complex DNA:protein interactions involved in different DNA processing mechanisms.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Anne Early, Lucy Drury, and Max Douglas for providing yeast strains for the overexpression of unlabeled proteins, as well as N.D. lab members Anuj Kumar, Katinka Ligthart, and Julien Gros for their help purifying loading factors and DDK. The authors also thank Kaley McCluskey, Dorian Mikolajczak, Joseph Yeeles, Jacob Lewis, Alessandro Costa, Hasan Yardimci, and Taekjip Ha for useful scientific discussions. D.R.M. acknowledges funding from a Boehringer Ingelheim Fonds PhD Fellowship. N.D. acknowledges funding from the Netherlands Organisation for Scientific Research (NWO) through Top grant 714.017.002 and Spinozapremie SPI 81-772, and from the European Research Council through an Advanced Grant (REPLICHROMA; grant number 789267).
Materials
| Name | Company | Catalog Number | Comments |
| AflII | NEB | R0520L | |
| Anti-digoxigenin coated polystyrene beads | Spheroteck | DIGP-20-2 | |
| ATP solution | Thermo Fisher | R0441 | |
| ATPγS | Roche | 11162306001 | |
| BSA | NEB | B9000S | |
| C-Trap | Lumicks | ||
| CutSmart Buffer | NEB | B6004S | Provided with AflII |
| dCTP | Promega | U122B | |
| D-Desthiobiotin-7-dATP | Jena Bioscience | NU-835-Desthiobio | |
| dGTP | Thermo Fisher | 10218014 | |
| Digoxigenin-11-dUTP | Jena Bioscience | NU-803-DIGXL | |
| Dynabeads M-280 Streptavidin magnetic beads | Invitrogen | 11205D | |
| Klenow Fragment (3'→5' exo-) | NEB | M0212L | |
| Microspin S-400 HR spin columns | GE Healthcare | GE27-5140-01 | |
| NEBuffer2 | NEB | B7002S | Provided with Klenow Fragment |
| Nonidet P 40 Substitute | Sigma | 74385 | |
| Pluronic F-127 | Sigma | P2443 |
References
- Bell, S. P., Labib, K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 203 (3), 1027-1067 (2016).
- Burnham, D. R., Kose, H. B., Hoyle, R. B., Yardimci, H. The mechanism of DNA unwinding by the eukaryotic replicative helicase. Nat Commun. 10 (1), 1-14 (2019).
- Ticau, S., Friedman, L. J., Ivica, N. A., Gelles, J., Bell, S. P. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell. 161 (3), 513-525 (2015).
- Ticau, S., et al. Mechanism and timing of Mcm2-7 ring closure during DNA replication origin licensing. Nat Str Mol Biol. 24 (3), 309-315 (2017).
- Gupta, S., Friedman, L. J., Gelles, J., Bell, S. P. A helicase- tethered ORC flip enables bidirectional helicase loading. eLife. 10, e74282 (2021).
- Lewis, J. S., et al. Single-molecule visualization of Saccharomyces cerevisiae leading-strand synthesis reveals dynamic interaction between MTC and the replisome. Proc Natl Acad Sci U S A. 114 (40), 10630-10635 (2017).
- Dulin, D., Berghuis, B. A., Depken, M., Dekker, N. H. Untangling reaction pathways through modern approaches to high-throughput single-molecule force-spectroscopy experiments. Curr Opinion Str Biol. 34, 116-122 (2015).
- Lewis, J. S., van Oijen, A. M., Spenkelink, L. M. Embracing heterogeneity: Challenging the paradigm of replisomes as deterministic machines. Chem Rev. 123 (23), 13419-13440 (2023).
- Abbondanzieri, E. A., Greenleaf, W. J., Shaevitz, J. W., Landick, R., Block, S. M. Direct observation of base-pair stepping by RNA polymerase. Nature. 438 (7067), 460-465 (2005).
- Yeeles, J. T. P., Deegan, T. D., Janska, A., Early, A., Diffley, J. F. X. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 519 (7544), 431-435 (2015).
- Douglas, M. E., Ali, F. A., Costa, A., Diffley, J. F. X. The mechanism of eukaryotic CMG helicase activation. Nature. 555 (7695), 265-268 (2018).
- Remus, D., et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 139 (4), 719-730 (2009).
- Frigola, J., Remus, D., Mehanna, A., Diffley, J. F. X. ATPase-dependent quality control of DNA replication origin licensing. Nature. 495 (7441), 339-343 (2013).
- Coster, G., Frigola, J., Beuron, F., Morris, E. P., Diffley, J. F. X. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell. 55 (5), 666-677 (2014).
- Coster, G., Diffley, J. F. X. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science. 357 (6348), 314-318 (2017).
- Miller, T. C. R., Locke, J., Greiwe, J. F., Diffley, J. F. X., Costa, A. Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature. 575 (7784), 704-710 (2019).
- Abid Ali, F., et al. Cryo-EM structure of a licensed DNA replication origin. Nat Commun. 8 (1), 1-10 (2017).
- Sánchez, H., et al. DNA replication origins retain mobile licensing proteins. Nat Commun. 12 (1), 1908 (2021).
- Ilves, I., Petojevic, T., Pesavento, J. J., Botchan, M. R. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 37 (2), 247-258 (2010).
- Moyer, S. E., Lewis, P. W., Botchan, M. R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 103 (27), 10236-10241 (2006).
- Langston, L. D., et al. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci U S A. 111 (43), 15390-15395 (2014).
- Sheu, Y. J., Stillman, B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 24 (1), 101-113 (2006).
- Sheu, Y. J., Stillman, B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 463 (7277), 113-117 (2010).
- Randell, J. C. W., et al. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 40 (3), 353-363 (2010).
- Greiwe, J. F., et al. Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase. Nat Str Mol Biol. 29 (1), 10-20 (2021).
- de Jesús-Kim, L., Friedman, L. J., Ramsoomair, C., Gelles, J., Bell, S. P. DDK regulates replication initiation by controlling the multiplicity of Cdc45-GINS binding to Mcm2-7. eLife. 10, 65471 (2021).
- Lewis, J. S., et al. Mechanism of replication origin melting nucleated by CMG helicase assembly. Nature. 606 (7916), 1007-1014 (2022).
- Ramírez Montero, D., et al. Nucleotide binding halts diffusion of the eukaryotic replicative helicase during activation. Nat Commun. 14 (1), 2082 (2023).
- Kose, H. B., Larsen, N. B., Duxin, J. P., Yardimci, H. Dynamics of the eukaryotic replicative helicase at lagging-strand protein barriers support the steric exclusion model. Cell Rep. 26 (8), 2113-2125 (2019).
- Lewis, J. S., et al. Single-molecule visualization of Saccharomyces cerevisiae leading-strand synthesis reveals dynamic interaction between MTC and the replisome. Proc Natl Acad Sci U S A. 114 (40), 10630-10635 (2017).
- Lewis, J. S., et al. Tunability of DNA polymerase stability during eukaryotic DNA replication. Mol Cell. 77, 1-9 (2020).
- Schauer, G. D., et al. Replisome bypass of a protein-based R-loop block by Pif1. Proc Natl Acad Sci U S A. 117 (48), 30354-30361 (2020).
- Wasserman, M. R., Schauer, G. D., O'Donnell, M. E., Liu, S. Replication fork activation is enabled by a single-stranded DNA gate in CMG helicase. Cell. 178 (3), 600-611 (2019).
- Kose, H. B., Xie, S., Cameron, G., Strycharska, M. S., Yardimci, H. Duplex DNA engagement and RPA oppositely regulate the DNA-unwinding rate of CMG helicase. Nat Commun. 11 (1), 1-15 (2020).
- White, D. S., Smith, M. A., Chanda, B., Goldsmith, R. H. Strategies for overcoming the single-molecule concentration barrier. ACS Meas Sci Au. 3 (4), 239-257 (2023).
- Liachko, I., et al. A comprehensive genome-wide map of autonomously replicating sequences in a naive genome. PLoS Genet. 6 (5), 22 (2010).
- Kapadia, N., et al. Processive activity of replicative DNA polymerases in the replisome of live eukaryotic cells. Mol Cell. 80 (1), 114-126 (2020).
- Claussin, C., Vazquez, J., Whitehouse, I. Single-molecule mapping of replisome progression. Mol Cell. 82 (7), 1372-1382 (2022).
- Polo Rivera, C., Deegan, T. D. Replicon-seq: seeing is believing. Trends Genet. 38 (10), 987-988 (2022).
- Sparks, J. L., et al. The CMG helicase bypasses DNA-protein cross-links to facilitate their repair. Cell. 176 (1-2), 167-181 (2019).
- Vrtis, K. B., et al. Single-strand DNA breaks cause replisome disassembly. Mol Cell. 81 (6), 1309-1318 (2021).
- Low, E., Chistol, G., Zaher, M. S., Kochenova, O. V., Walter, J. C. The DNA replication fork suppresses CMG unloading from chromatin before termination. Genes Dev. 34 (21), 1534-1545 (2020).
- Parker, M. W., et al. A new class of disordered elements controls DNA replication through initiator self-assembly. eLife. 8, 1-35 (2019).
- Hirsch, J. D., et al. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: Uses for protein labeling, detection, and isolation. Anal Biochem. 308 (2), 343-357 (2002).
- Candelli, A., Wuite, G. J. L., Peterman, E. J. G. Combining optical trapping, fluorescence microscopy and micro-fluidics for single molecule studies of DNA-protein interactions. Phys Chem Chem Phys. 13 (16), 7263-7272 (2011).
- Candelli, A., et al. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc Natl Acad Sci U S A. 111 (42), 15090-15095 (2014).
- Bustamante, C., Marko, J. F., Siggia, E. D., Smith, S. Entropic elasticity of lambda-phage DNA. Science. 265 (5178), 1599-1600 (1994).
- Liu, Z., et al. A biophysics toolbox for reliable data acquisition and processing in integrated force-confocal fluorescence microscopy. ACS Photonics. 11 (4), 1592-1603 (2024).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved