斑块测定:确定病毒性皮石为斑块形成单位(PFU)的方法
Overview
资料来源:蒂尔德·安徒森1号,罗尔夫·卢德1
1临床科学隆德系,感染医学系,生物医学中心,隆德大学,221 00 隆德,瑞典
感染原核生物的病毒,称为噬菌体或简称噬菌体,在20世纪初由Twort(1)和d'Hérelle(2)独立发现。自那时以来,噬菌体因其治疗价值(3)及其对人类(4)以及全球生态系统(5)的影响而得到广泛认可。目前的关切促使人们重新关注使用噬菌体作为现代抗生素替代治疗传染病的方法(6)。基本上,所有的噬菌体研究都依赖于纯化和量化病毒的能力,也称为病毒性小子。最初描述在1952年,这是斑块测定的目的(7)。几十年后,多项技术进步,斑块测定仍然是确定病毒性牙点(8)的最可靠的方法之一。
噬菌体通过将遗传物质注入宿主细胞,劫持机器来生产新的噬菌体颗粒,并最终导致宿主通过细胞解释释放许多后代病毒。由于其微小的大小,噬菌体不能只用光显微镜观察;因此,扫描电子显微镜是必需的(图1)。此外,噬菌体不能像细菌一样在营养琼脂板上培养,因为它们需要宿主细胞来捕食。
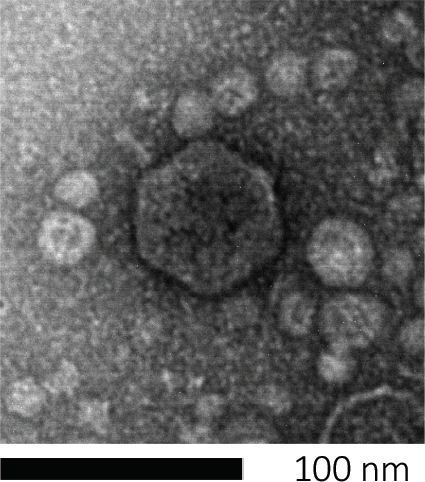
图1:噬菌体形态,这里以大肠杆菌噬菌体为例子,可以用扫描电子显微镜进行研究。大多数噬菌体属于考多病毒(尾噬菌体)。这种特殊的噬菌体有一个很短的尾巴结构和一个食道头,把它放在波多病毒家族。
斑块测定(图2)基于将宿主细胞(优先用于对数相生长)并入培养基。这创造了一个密集,浑浊的细菌层,能够维持病毒生长。分离的噬菌体随后可以感染、在一个细胞内复制和分莱。每个分莱细胞,多个相邻的细胞立即被感染。在几个周期中,在原本浑浊的板块中可以观察到一个清晰的区域(斑块),表明最初是单个噬菌体粒子的存在。因此,样品每体积(即PFU/mL)的斑块形成单位数量可以从产生的斑块数量中确定。
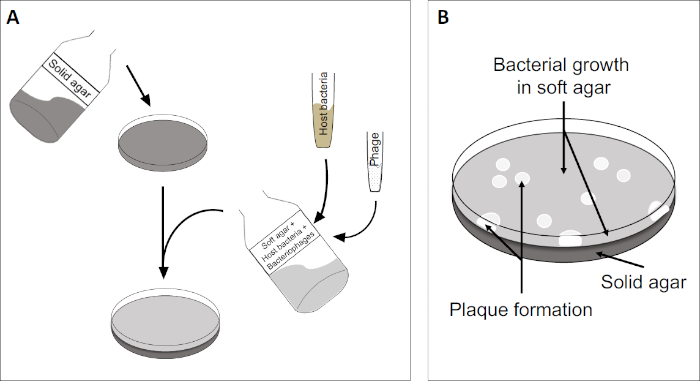
图2:检测斑块形成单元(PFU)是确定样品中噬菌体数量的常见方法。(A)无菌培养皿的基底覆盖着适当的固体营养介质,然后是软培养基、易感宿主细胞的混合物和原噬菌体样本的稀释。请注意,在某些情况下,噬菌体悬浮液也可以均匀地分布在已经凝固的软琼脂表面。(B)宿主细菌的生长在琼脂层形成细胞草坪。噬菌体复制产生由宿主细胞解致引起的透明区域或斑块。
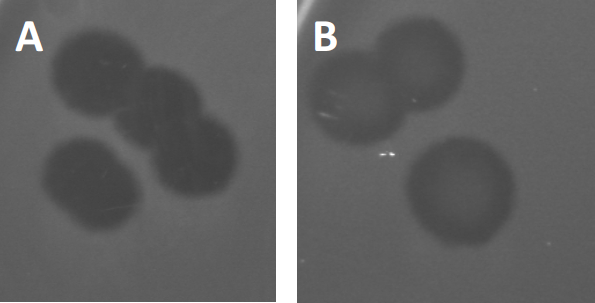
图3:PFU测试结果显示,噬菌体产生的多斑块。由于易感宿主细胞的赖清,斑块可被视为细菌草坪上的清除区,要么具有(A)完全清除,要么(B)由产生耐药细菌(或可能是由耐温噬菌体)引起的部分再生致源循环)。
除了以前描述的溶血生长外,某些温带噬菌体可以采用所谓的溶源性生命周期。在地合体中,病毒通过将其遗传物质纳入宿主细胞(9)的基因组而假定为潜伏状态,这通常对进一步的噬菌体感染具有抵抗力。这有时通过斑块的轻微云彩(图3B)来揭示。然而,值得注意的是,斑块也可能显得模糊,由于细菌的再生长,已经进化了抵抗噬菌体,独立于以前的噬菌体感染。
病毒可以附着或吸附,只有有限的宿主细菌(10)。宿主范围受到细胞内抗病毒策略(如CRISPR-Cas系统(11)的进一步限制。细菌亚群对特定噬菌体表现出的抗药性/敏感性历来被用来将细菌菌株分类为不同的噬菌体类型(图4)。虽然这种方法的有效性现已被新的测序技术所超越,但噬菌体类型仍然可以提供有关细菌-噬菌体相互作用的宝贵信息,例如,促进为临床使用设计噬菌体鸡尾酒.
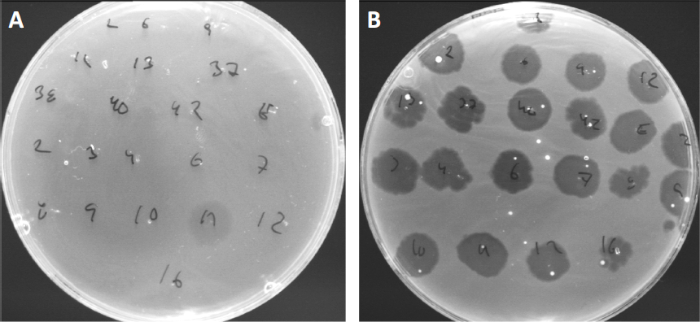
图4:不同细菌菌株的噬菌体敏感性。软琼脂板与可爱痤疮菌株(A)AD27和(B)AD35,被发现与21种不同的C.痤疮噬菌体。只有噬菌体11能够感染和杀死AD27,而AD35菌株对所有噬菌体表现出敏感性。这种技术,称为噬菌体类型,可用于根据噬菌体易感性将细菌种类和菌株分成不同的亚群。
Procedure
1. 设置
- 在开始任何涉及微生物的工作之前,请确保工作空间已消毒(例如,用 70% 乙醇擦拭)。始终穿着实验室外套和手套,保持长发绑在背上,并确保任何伤口都特别好保护。
- 完成后,对所有表面进行消毒,彻底清洗/消毒手和手腕。
2. 议定书
- LB 介质准备
注意:根据宿主细菌菌株和噬菌体的不同,不同的液体介质可能更适合于宿主细菌菌株的初始培养,或者不同的固体介质可能更适合后续生长。莱索根汤 (LB) 用于此协议中的肉汤和琼脂。- 混合 4 g LB 在 200 mL 蒸馏水中,在三重,为 LB 肉汤, 固体底部琼脂, 和软顶部琼脂.所有解决方案都应在容器中制备,其容量应为最终体积的两倍,以防止高压灭菌时溢出。
注意:如果以三重分法执行测定,则准备将 LB 底部琼脂的量翻倍。 - 根据需要
- 混合 4 g LB 在 200 mL 蒸馏水中,在三重,为 LB 肉汤, 固体底部琼脂, 和软顶部琼脂.所有解决方案都应在容器中制备,其容量应为最终体积的两倍,以防止高压灭菌时溢出。
Application and Summary
References
- Twort, F. An investigation on the nature of ultra-microscopic viruses. Lancet. 186 (4814): 1241-1243. (1915)
- d'Hérelle, F. An invisible antagonist microbe of dysentery bacillus. Comptes Rendus Hebdomadaires Des Seances De L Academie Des Sciences. 165: 373-375. (1917)
- Cisek AA, Dąbrowska I, Gregorczyk KP, Wyżewski Z. Phage Therapy in Bacterial Infections Treatment: One Hundred Years After the Discovery of Bacteriophages. Current Microbiology. 74 (2):277-283. (2017)
- Mirzaei MK, Maurice CF. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nature Reviews Microbiology. 15 (7):397. (2017)
- Breitbart M, Bonnain C, Malki K, Sawaya NA. Phage puppet masters of the marine microbial realm. Nature Microbiology. 3 (7):754-766. (2018)
- Leung CY, Weitz JS. Modeling the synergistic elimination of bacteria by phage and the innate immune system. Journal of Theoretical Biology. 429:241-252. (2017)
- Dulbecco R. Production of Plaques in Monolayer Tissue Cultures by Single Particles of an Animal Virus. Proceedings of the National Academy of Sciences of the United States of America. 38 (8):747-752. (1952)
- Juarez D, Long KC, Aguilar P, Kochel TJ, Halsey ES. Assessment of plaque assay methods for alphaviruses. J Virol Methods. 187 (1):185-9. (2013)
- Clokie MRJ, Millard AD, Letarov AV, Heaphy S. 2011. Phages in nature. Bacteriophage. 1 (1):31-45. (2011)
- Moldovan R, Chapman-McQuiston E, Wu XL. On kinetics of phage adsorption. Biophys J. 93 (1):303-15. (2007)
- Garneau JE, Dupuis M-È, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S.. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 468 (7320):67. (2010)
Tags
跳至...
此集合中的视频:

Now Playing
斑块测定:确定病毒性皮石为斑块形成单位(PFU)的方法
Microbiology
185.6K Views

创建维诺格拉茨基柱:在沉积物样本中丰富微生物物种的方法
Microbiology
128.2K Views

连续稀释和电镀:微生物枚举
Microbiology
313.9K Views

富集培养:在选择性和微分介质上培养有氧和厌氧微生物
Microbiology
131.7K Views

纯培养和条纹电镀:从混合样品中分离单一细菌菌落
Microbiology
165.7K Views

16S rRNA测序:基于PCR的技术,用于识别细菌物种
Microbiology
187.9K Views

生长曲线:使用菌落成形单位和光学密度测量生成生长曲线
Microbiology
293.0K Views

抗生素敏感性测试:Epsilo计测试,以确定两种抗生素的MIC值,并评估抗生素协同效应
Microbiology
93.4K Views

显微镜和染色:革兰克、胶囊和内孔染色
Microbiology
362.4K Views

使用自适应氯化钙程序对大肠杆菌细胞的转化
Microbiology
86.3K Views

结合:将阿霉素耐药性从捐赠者转移到受体大肠杆菌的方法
Microbiology
38.1K Views

噬菌体转导:将氨基青霉素耐药性从捐赠者转移到受体大肠杆菌的方法
Microbiology
28.9K Views
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。
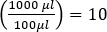 即)
即)