A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Spatial High-resolution Analysis of Gene Expression Levels in Tendons
In This Article
Summary
This article describes how to perform an optimized in situ protocol for tendons. This method discusses tissue preparation, section permeabilization, probe design, and signal amplification methods.
Abstract
In recent years, many protocols have been developed for high-resolution transcriptomics in many different medical and biology fields. However, matrix-rich tissues, and specifically, tendons were left behind due to their low cell number, low RNA amount per cell, and high matrix content, which made them complicated to analyze. One of the recent and most important single-cell tools is the spatial analysis of gene expression levels in tendons. These RNA spatial tools have specifically high importance in tendons to locate specific cells of new and unknown populations, validate single-cell RNA-seq results, and add histological context to the single-cell RNA-seq data. These new methods will enable the analysis of RNA in cells with exceptional sensitivity and the detection of single-molecule RNA targets at the single-cell level, which will help to molecularly characterize tendons and promote tendon research.
In this method paper, we will focus on the available methods to analyze spatial gene expression levels on histological sections by using novel in situ hybridization assays to detect target RNA within intact cells at single-cell levels. First, we will focus on how to prepare the tendon tissue for the different available assays and how to amplify target-specific signals without background noise but with high sensitivity and high specificity. Then, the paper will describe specific permeabilization methods, the different probe designs, and the signal amplification strategies currently available. These unique methods of analyzing transcription levels of different genes in single-cell resolution will enable the identification and characterization of the tendon tissue cells in young and aged populations of various animal models and human tendon tissues. This method will also help analyze gene expression levels in other matrix-rich tissues such as bones, cartilage, and ligaments.
Introduction
Tendons are connective tissues that enable the transmission of force between muscle and bone1. Developmentally, axial tenocytes are derived from mesenchymal cells within the sclerotome of the somites2; limb tendons derive from the lateral plate mesoderm; and cranial tendons arise from the cranial neural crest lineage3,4. Tendon can be characterized by the expression of the scleraxis transcription factor5, although several markers also play a key role in tendon development, including tenomodulin, mohawk, and early growth response 1/26,7,8,9.
Despite the few known markers of the tendon, in general, a more in-depth characterization remains challenging because the tendon contains cells that span across a gradient of biomechanical properties. From the myotendinous junction, tendon mid-body, and the more calcified enthesis, the tendon cells reside in extracellular matrices that range in tensile properties. Since the tendon must withstand tensile stress imposed by the difference in mechanical strength between soft and hard tissue, the spatial organization of the cells in the tendon is particularly important for its function. However, little is known about these tendon subpopulations.
Many high-resolution spatial transcriptomic tools can be used to begin to elucidate cell subpopulations, including but not limited to, single-cell RNA Seq or in situ hybridization. However, while these spatial profiling assays help uncover RNA expression across tissue after microdissection or sectioning, these methods can be challenging when performed on tendon tissue. Tendons are matrix-rich tissues composed of nearly 86% of collagen by dry mass10, making it challenging to extract the cells for sequencing. Due to both the complications in isolating cells from the matrix, the hypocellular nature of the tendon11, and the relatively low RNA count, the tendon is a difficult tissue to analyze.
In this paper, we present a method to optimize novel in situ hybridization assays to leverage them for tendons by providing tissue preparation, permeabilization, and probe design methods. Coupled with existing sequencing technologies, this may help researchers spatially characterize tendon subpopulations across developing, adult, or injured tendons with increased assay sensitivity and specificity.
Protocol
All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) and AAALAC guidelines. Experiments were performed under approved protocol #2013N000062 at Massachusetts General Hospital. In this study, C57BL/J6 mice (5 weeks of age and P0) were used. See the Table of Materials for details related to all materials, reagents, and instruments used in this protocol.
1. Sample preparation and fixation
- Euthanize mice in a CO2 chamber followed by cervical dislocation as a secondary method of euthanasia. Then, use scissors to cut the mouse hindlimbs along the hip joint12. In a scintillation vial, immerse the mouse hindlimb in enough 4% paraformaldehyde (PFA) solution to completely cover the sample, and fix the tissue for 24 h.
NOTE: Instead of 4% PFA, 10% formalin may also be used for fixation. After fixation, the sample may be stored in phosphate-buffered saline (PBS) until further processing. By fixing the whole hindlimb, we are able to preserve tension on the tendon and have the opportunity to examine the enthesis, tendon midbody, or myotendinous junction. - After removing the PFA from the scintillation vial, wash the sample for 3 x 10 min with 1x PBS by adding enough PBS to cover the sample. Then, immerse the hindlimbs in 0.5 M EDTA at 4 °C with agitation for 1-2 weeks, replacing with fresh EDTA every 2-3 days.
- After removing the EDTA, wash the sample for 3 x 10 min with 1x PBS, dehydrate the samples in 70% EtOH, and embed them in paraffin in the desired orientation. Use conventional FFPE sectioning techniques to section the tissue with a thickness of 3 µm and transfer the sections onto a treated, charged microscope slide (see Table of Materials)13.
NOTE: Sections can be stored in 4 °C prior to proceeding to the next steps. - Deparaffinization
- Bake the slides on a hot plate at 60 °C for 1 h and then allow the slides to come to room temperature (RT). To remove the paraffin wax from the slides, place the slides on a slide rack and lower them into a staining dish filled with xylene. Repeat in fresh xylene for 3 x 5 min.
CAUTION: Xylene is hazardous; handle with care. - To rehydrate the sections, immerse the slides in the following for 5 min per wash, all at RT: wash 2x in 100% EtOH, 1x in 75% EtOH diluted in dH2O, 1x in 50% EtOH diluted in PBS, 1x in 25% EtOH diluted in PBS, and 2x in PBS.
NOTE: All the rehydration steps are done with the slides on a slide rack and with the various EtOH solutions in staining dishes.
- Bake the slides on a hot plate at 60 °C for 1 h and then allow the slides to come to room temperature (RT). To remove the paraffin wax from the slides, place the slides on a slide rack and lower them into a staining dish filled with xylene. Repeat in fresh xylene for 3 x 5 min.
2. RNAscope (commercialized ISH) protocol14 adaptation
- Preparing solutions and probes
- Prepare TEG solution for the pretreatment of sectioned samples by combining 25 mM of Tris-HCl at pH 8, 10 mM of EDTA, and 50 mM of glucose. Make a sufficient volume for all samples to be treated or prepare excess; filter-sterilize and store at RT; mix the solution properly before using it.
- Prepare enough 1x Wash Buffer (see the Table of Materials) for all washes and store it at RT.
- In a 40 °C water bath or incubator, warm the probes of interest for 10 min. Then, prepare the probes according to the ISH protocol14.
- Pretreatment of samples
- Rather than using the recommended pretreatment solution, immerse the slides in a staining dish containing TEG buffer for 4 h at 60 °C.
NOTE: TEG buffer incubation can be extended for up to 6 h if the tissue is particularly dense or if antigen retrieval of the RNA binding motif of interest is challenging. In that case, a stop point can be added here, the slides dried and kept in a 4 °C fridge overnight. - Remove the samples from the buffer and let them dry. Then, draw around the samples with a hydrophobic barrier pen, immerse them in another staining dish containing protease IV, and incubate them in an oven for 45 min at 40 °C.
NOTE: We recommend protease IV for adult mice. If using younger mice, such as P0, we recommend using a milder enzyme, such as protease III, for 30 min. When troubleshooting for different ages, observe the integrity of the tendon and check for degradation. If the tissue is dissociating or degrading, decrease the incubation time or change the protease used. - Wash samples again in TEG buffer for 30 min and then hybridize the probes according to the ISH protocol14.
- Add 50-100 µL of mounting reagent on top of the sample. Place a coverslip on top of the samples for microscopy. When imaging, use a high magnification (40x objective or higher) to view the signal.
- Rather than using the recommended pretreatment solution, immerse the slides in a staining dish containing TEG buffer for 4 h at 60 °C.
3. HCR ISH protocol15 adaptation
- Postfixation
- Prepare 4% PFA (in PBS), PBT solution (0.1% Tween-20 in PBS), and Proteinase K in PBS (stock concentration of 10 mg/mL, final concentration of 5 µg/mL).
- To begin the first postfix step, immerse the slides in a staining dish containing 4% PFA for 5 min; then, remove the 4% PFA solution and store it for use in later steps. Rinse the slides for 2 x 5 min in PBT solution and remove the PBT solution. Place the slides in a staining dish containing Proteinase K solution (5 µg/mL) for 5 min and rinse them in the PBT solution 2 x 5 min.
- Begin the second postfix step and immerse the slides in the 4% PFA for 5 min.
NOTE: This is the same 4% PFA saved from step 3.1.2. - Rinse the slides for 3 x 5 min in the PBT solution.
NOTE: All the postfixation steps are done with the slides on a slide rack and in staining dishes containing the respective solutions.
- Acetylation
- Prepare the acetylation solution containing 625 µL of acetic anhydride, 3.3 mL of 1 M triethanolamine (TEA) buffer, and 246 mL of ddH2O. Be sure to mix well and use immediately after preparation.
- Place the slides in the acetylation solution for 10 minutes and then rinse 3 x 5 min in PBT solution. Rinse the slides in ddH2O and let them air dry for 30 min. Then, draw around the samples with a hydrophobic barrier pen.
NOTE: All the acetylation steps are done with the slides on a slide rack and in staining dishes containing the respective solutions
- Hybridization
- Preheat the hybridization buffer to 37 °C and prewarm a humidified chamber to 37 °C.
CAUTION: The buffer contains formamide, a hazardous material. If the slides are not sufficiently dry, blot the edges with a laboratory wipe to remove excess dH2O. - Prepare probe solutions by adding 0.4 pmol of each probe mixture to 100 µL of hybridization buffer.
NOTE: If using dHCR imaging, use a higher concentration of probe to improve probe hybridization efficiency. - Prepare 4 L of Sodium Chloride Sodium Citrate Buffer (20x SSC) by combining 3 M NaCl (701.1 g of NaCl in a final volume of 4 L) and 0.3 M of Na3CH6H5O7•2H2O (352.8 g) and then adjusting the final pH to 7 by adding HCl or 10 N NaOH. Add enough H2O to reach 4 L. Prepare 5x SSCT by diluting 20x SSC to 5x and adding enough 10% Tween 20 so that it is 0.1% of the total volume.
- Add 200 µL of hybridization buffer to the sample, place the slides in a humidified chamber, and let the slides incubate for 10 min. Remove the hybridization solution and drain the excess buffer on the slide by blotting the edges with a laboratory wipe.
- Add 100 µL of the prepared probe solution on top of the sample and place a coverslip on the sample. Incubate in an oven overnight or for around 12-16 h in a chamber humidified with a solution containing 5x SSC and 50% formamide. Set the temperature to 37 °C.
NOTE: Since the incubation is overnight, the addition of the coverslip is to minimize evaporation. - Leave an aliquot of 5x SSCT solution in a 37 °C water bath and a second aliquot at RT. Anticipate the volumes of these aliquots needed based on the sample number and the number of washes in the next step.
- Preheat the hybridization buffer to 37 °C and prewarm a humidified chamber to 37 °C.
- Washing
- Using the previously preheated 5x SSCT solution, prepare 75% wash buffer/25% 5x SSCT, 50% wash buffer/50% 5x SSCT, and 25% wash buffer/75% 5x SSCT solutions.
- To remove excess probes, immerse the slides in series for 15 min per wash at 37 °C in 75% wash buffer/25% 5x SSCT, 50% wash buffer/50% 5x SSCT, and 25% wash buffer/75% 5x SSCT. Allow the coverslips to float off from the samples.
- Incubate the slides 2 x 15 min with 100% 5x SSCT and then immerse them in 5x SSCT for 5 min at RT.
NOTE: The washing steps can be done with the slides on a slide rack and in staining dishes containing the respective solutions or with the solutions added as droplets.
- Amplification
- Prepare 6 pmol of hairpin h1 and 6 pmol of hairpin h2 by snap cooling 2 µL of a 3 µM stock (heat at 95 °C for 90 s and cool to RT). Protect the hairpins from light during this process.
NOTE: HCR hairpins h1 and h2 are provided in a hairpin storage buffer and are ready for snap cooling. Snap cool h1 and h2 hairpins in separate tubes. - Dry the slides by blotting their edges with a laboratory wipe. Then, add 200 µL of amplification buffer on top of the sample and place it in a humidified chamber for 30 min at RT. Prepare the hairpin mixture by adding snapcooled h1 hairpins and snapcooled h2 hairpins to 100 µL of amplification buffer at RT.
- Remove the amplification buffer and drain the excess liquid on the slide by blotting the edges with a laboratory wipe. Add 100 µL of the previously prepared hairpin mixture and place parafilm on top of the sample. Incubate the slides for a minimum of 4 h or overnight in a dark, humidified chamber at RT.
- To remove excess hairpins, immerse the slides in 5x SSCT in a staining dish and incubate at RT for 30 min. Repeat the wash in fresh 5x SSCT for 30 min, and a third time for 5 min.
- Dry the slides by blotting the edges with a laboratory wipe and add 50-100 µL of mounting reagent on top of the sample. Place a coverslip on top of the samples for microscopy. When imaging, use a high magnification (40x objective or higher) to view the signal.
- Prepare 6 pmol of hairpin h1 and 6 pmol of hairpin h2 by snap cooling 2 µL of a 3 µM stock (heat at 95 °C for 90 s and cool to RT). Protect the hairpins from light during this process.
Results
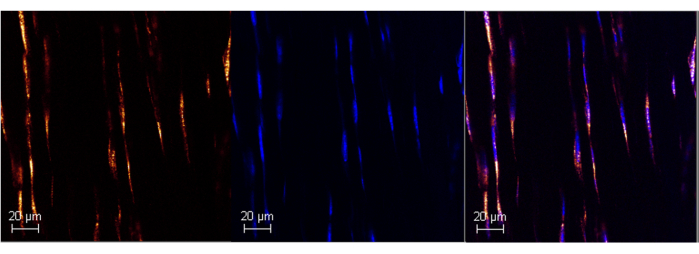
Figure 1: Poly A RNA expression in adult mouse Achilles tendon using RNAScope. Representative image of successful Poly A labeling in mouse Achilles tendon (left panel) using the commercialized ISH assay. Colocalization with DAPI confirms the specificity of the probe (middle and right panels), allowing control for background noise. Images were t...
Discussion
In this paper, we describe modifications made to leverage existing ISH tools such that they can be used in tendon tissue with a high degree of specificity and sensitivity. Since the tendon is a highly matrix-dense tissue, protocol adjustments must often be made to achieve similar degrees of probe penetration and specificity. These specific permeabilization methods and signal amplification strategies of the tendon tissue are integral to improving the efficacy of the ISH protocols discussed. Without these steps, it is chal...
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
The authors thank Jenna Galloway and the members of Galloway Lab for their support and encouragement in the development and troubleshooting of these protocols.
Materials
| Name | Company | Catalog Number | Comments |
| 1 M triethanolamine buffer | |||
| 10% Formalin solution | |||
| 10% Tween-20 | |||
| 20x Saline Sodium Citrate buffer | |||
| 4% PFA | |||
| ACD RNAscope Fluorescent Multiplex Fluorescent Reagent Kit V2 | ACD | 323100 | |
| Acetic Anhydride | |||
| Axio Imager Microscope | ZEISS | ||
| C57BL/J6 mice | JAX ID: 000664 | ||
| Coverslips | Fisher | 12-541-042 | |
| ddH2O | |||
| ETDA | Thermofisher | AM9262 | |
| EtOH | |||
| Glucose | VWR Chemicals BDH | BDH9230-500G | |
| HCR RNA-FISH Bundle | Molecular Instruments Inc. | ||
| HybEZ II Hybridization System | ACD | ||
| Immedge Barrier Pen | Vector Laboratories | H4000 | |
| Leica SPE Confocal Microscope | Leica | ||
| Parafilm | Fisher | ||
| Phosphate-buffered saline (PBS, 1x) | Invitrogen | AM9625 | Dilute 10x PBS in milli-Q water to get 1x solution |
| Protease IV | |||
| Proteinase K | Roche | 3115836001 | |
| RNAscope H2O2 and Protease Reagents | ACD | PN 322381 | Included in ACD RNAscope Fluorescent Multiplex Fluorescent Reagent Kit V3 |
| RNAscope Multiplex Fluorescent Detection Kit | ACD | PN 323110 | Included in ACD RNAscope Fluorescent Multiplex Fluorescent Reagent Kit V2 |
| RNAscope Target Retrieval reagents | ACD | 322000 | Included in ACD RNAscope Fluorescent Multiplex Fluorescent Reagent Kit V4 |
| RNAscope Wash Buffer | ACD | PN 310091 | Included in ACD RNAscope Fluorescent Multiplex Fluorescent Reagent Kit V5 |
| RNAscope Probe Diluent | ACD | 300041 | |
| Slide holder | StatLab | 4465A | |
| Staining Dish with Lid | StatLab | LWS20WH | |
| Superfrost Plus Microscope slides | Fisher | 1255015 | treated, charged slides |
| Tris-HCl | |||
| Xylene | Sigma-Aldrich | 534056-4L |
References
- Sharma, P., Maffulli, N. Tendon injury and tendinopathy: healing and repair. Journal of Bone and Joint Surgery. 87 (1), 187-202 (2005).
- Brent, A. E., Schweitzer, R., Tabin, C. J. A somitic compartment of tendon progenitors. Cell. 113 (2), 235-248 (2003).
- Noden, D. M. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Developmental Biology. 96 (1), 144-165 (1983).
- Chen, J. W., Galloway, J. L. The development of zebrafish tendon and ligament progenitors. Development. 141 (10), 2035-2045 (2014).
- Schweitzer, R., et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 128 (19), 3855-3866 (2001).
- Tsai, S., Nödl, M., Galloway, J. Bringing tendon biology to heel: Leveraging mechanisms of tendon development, healing, and regeneration to advance therapeutic strategies. Developmental Dynamics. 250 (3), 393-413 (2021).
- Kannus, P., et al. Location and distribution of non-collagenous matrix proteins in musculoskeletal tissues of rat. The Histochemical Journal. 30 (11), 799-810 (1998).
- Kannus, P. Structure of the tendon connective tissue. Scandinavian Journal of Medicine & Science in Sports. 10 (6), 312-320 (2000).
- Thorpe, C. T., Birch, H. L., Clegg, P. D., Screen, H. R. C. The role of the non-collagenous matrix in tendon function. International Journal of Experimental Pathology. 94 (4), 248-259 (2013).
- Lin, T. W., Cardenas, L., Soslowsky, L. J. Biomechanics of tendon injury and repair. Journal of Biomechanics. 37 (6), 865-877 (2004).
- Grinstein, M., et al. A distinct transition from cell growth to physiological homeostasis in the tendon. eLife. 8, e48689 (2019).
- Villaseñor, S., Grinstein, M. Two-photon microscopy for the study of tendons. Journal of Visualized Experiments. , e65853 (2023).
- Qin, C., et al. The cutting and floating method for paraffin-embedded tissue for sectioning. Journal of Visualized Experiments. (139), e58288 (2018).
- Wang, F., et al. RNAscope®: A Novel In Situ RNA Analysis Platform for Formalin-Fixed Paraffin-Embedded Tissues. Journal of Molecular Diagnostics. 14 (1), 22-29 (2012).
- Choi, H. M. T., et al. Third-generation situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 145 (12), 165753 (2018).
- Jackson, D. J., Herlitze, I., Hohagen, J. A whole mount in situ hybridization method for the gastropod mollusc Lymnaea stagnalis. Journal of Visualized Experiments. (109), e53968 (2016).
- Young, A. P., Jackson, D. J., Wyeth, R. C. A technical review and guide to RNA fluorescence in situ hybridization. PeerJ. 8, e8806 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved