Analyse du cycle cellulaire : utilisation de la coloration CFSE et de la cytométrie de flux pour évaluer la prolifération des lymphocytes T CD4 et CD8 après stimulation
Vue d'ensemble
Source: Perchet Thibaut1,2,3, Meunier Sylvain1,2,3, Sophie Novault4, Rachel Golub1,2,3
1 Unité de lymphpoiesis, Département d'immunologie, Institut Pasteur, Paris, France
2 INSERM U1223, Paris, France
3 Université Paris Diderot, Sorbonne Paris Cité, Cellule Pasteur, Paris, France
4 Flow Cytometry Platfrom, Cytometry and Biomarkers UtechS, Center for Translational Science, Institut Pasteur, Paris, France
Le cycle cellulaire est un processus universel de vie. Pendant le cycle cellulaire, une cellule subit plusieurs modifications pour se diviser en deux cellules filles. Ce mécanisme se produit tout au long de la vie d'un organisme en réponse à ses besoins. Les divisions cellulaires et le développement embryonnaire produisent un organisme complet à partir d'un zygote unicellulaire. À l'âge adulte, le cycle cellulaire est au cœur de nombreux processus biologiques critiques, tels que la réparation des tissus.
Les mécanismes de division cellulaire sont des événements étroitement contrôlés où la cellule subit des modifications progressives avant la division finale. Les cellules qui ne sont pas encore dans le cycle sont décrites comme étant dans la phase gap 0 (G0). Pendant cette étape, la cellule est considérée comme quiescente. Lorsque la cellule commence à cycle, quatre phases distinctes sont reconnues: Gap 1 (G1), Synthèse (S), Gap 2 (G2) et Mitose (M). La phase G1 est un point de contrôle des ressources nécessaires à la synthèse de l'ADN par la cellule. Ensuite, la phase S se produit, et la réplication de l'ADN commence, suivie par l'interphase G2, un autre point de contrôle qui contrôle tous les éléments nécessaires pour que la cellule se divise. Enfin, la cellule entre dans la mitose et se divise en deux cellules filles.
La division cellulaire est un paramètre très instructif dans de nombreux systèmes biologiques différents. Dans le domaine de l'immunologie, l'analyse de la prolifération des leucocytes peut indiquer le mécanisme de la réponse immunitaire. D'autres domaines d'investigation reposent également sur l'analyse du cycle cellulaire. Par exemple, l'analyse du cycle cellulaire pendant le développement de tumeur a amélioré notre compréhension du cancer.
De nombreux colorants fluorescents sont maintenant disponibles pour suivre la prolifération cellulaire. Ces colorants diffèrent dans leurs propriétés chimiques et spectrales. Deux classes différentes de colorants existent : les colorants protéiques se combinent en permanence avec les protéines en formant un lien covalent, et les colorants membranaires s'intercalent de façon stable dans les membranes cellulaires par l'intermédiaire de fortes associations hydrophobes. Les études in vitro et in vivo de la prolifération des cellules immunitaires par cytométrie du flux sont parmi les applications les plus courantes des deux classes de colorants de suivi cellulaire (1, 2).
CFSE (Carboxyfluorescein succinimidyl ester) est un colorant fluorescent qui marque les cellules qui divisent. Au départ, toutes les cellules reçoivent la même quantité de colorant; les cellules de division divisent uniformément le colorant qu'elles ont reçu entre leurs deux cellules de fille. Par conséquent, le cycle cellulaire peut être suivi par la diminution progressive de l'intensité des colorants dans les cellules. La coloration CFSE est suivie d'une cytométrie multiparamétrique conventionnelle, une technologie à haut débit à base de fluorescence qui permet une caractérisation phénotypique et fonctionnelle des cellules en fonction de leur degré de coloration CFSE (3).
Dans l'expérience suivante, nous évaluons la prolifération des cellules CD4et CD8et T in vitro,après stimulation CD3, en utilisant la cytométrie de coloration et de flux de cfSE.
Procédure
1. Préparation
- Avant de commencer, enfilez des gants de laboratoire et les vêtements de protection appropriés.
- Stériliser tous les outils de dissection, d'abord avec un détergent, puis avec 70% d'éthanol, puis les essuyer soigneusement.
- Préparer 50 ml de la solution de sel équilibrée de Hank (HBSS) contenant 2 % de sérum fœtal de veau (FCS).
2. Dissection
- À l'aide d'un système de distribution de dioxyde de carbone,
Résultats
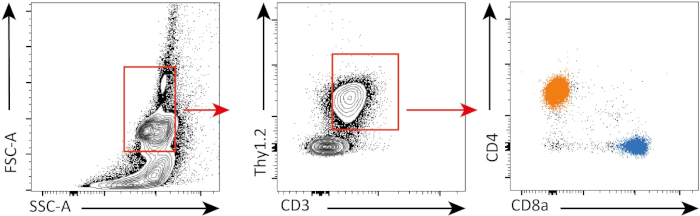
Dans cette expérience, nous avons suivi la prolifération des cellules Splénic CD4et CD8t dans la culture in vitro. Après 3 jours, nous n'avons pas vu la prolifération forte dans les deux CD4et CD8- cellules T avec ou sans stimulation. C'est ce que l'on peut voir sur le panneau supérieur de la figure 2 où les pics de CSFE ne diminuent pas. Cependant, après 5 jours, nous avons commencé à voir une pr...
Applications et Résumé
Les tests de prolifération sont souvent utilisés dans différents domaines tels que l'immunologie pour déterminer le degré d'activation des cellules. Il est également effectué dans le diagnostic d'oncologie pour déterminer l'agressivité de tumeur dans les patients. La coloration CFSE est une technique utile pour suivre la prolifération des populations de cellules immunitaires au fil du temps. D'autres méthodes permettent la caractérisation du cycle cellulaire. BrdU, un équivalent de CFSE est incorporé seulem...
References
- Lyons, A. B. and Parish, C. R. Determination of lymphocyte division by flow cytometry. Journal of Immunological Methods. 171 (1): 131-37, (1994).
- Lyons, A. B. Analyzing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. Journal of Immunological Methods. 243 (1-2), 147-154, (2000).
- Quah, B. J., Warren H. S., and Parish, C. R. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nature Protocols. 2 (9): 2049-56, (2007).
Tags
Passer à...
Vidéos de cette collection:

Now Playing
Analyse du cycle cellulaire : utilisation de la coloration CFSE et de la cytométrie de flux pour évaluer la prolifération des lymphocytes T CD4 et CD8 après stimulation
Immunology
24.2K Vues

Cytométrie en flux et tri cellulaire activé par fluorescence (FACS) : isolement des lymphocytes B spléniques
Immunology
92.9K Vues

Tri cellulaire magnétique (MACS) : isolement des lymphocytes T thymiques
Immunology
22.9K Vues

Tests ELISA : Indirect, en sandwich et par compétition
Immunology
238.1K Vues

Test ELISPOT : Détection des splénocytes sécrétants l'IFNgamma
Immunology
28.4K Vues

Immunohistochimie et immunocytochimie : Imagerie tissulaire par microscopie optique
Immunology
78.8K Vues

Génération d'anticorps monoclonaux à l'aide d'hybridomes
Immunology
43.5K Vues

Microscopie à fluorescence : coloration par immunofluorescence des sections de tissus inclus en paraffine
Immunology
53.8K Vues

Microscopie confocale à fluorescence : une technique pour localiser les protéines dans les fibroblastes de souris
Immunology
43.1K Vues

Techniques basées sur l'immunoprécipitation : purification des protéines endogènes à l'aide de billes d'agarose
Immunology
87.6K Vues

Transfert adoptif de cellules : introduction de splénocytes d'une souris donneuse vers une souris hôte et évaluation du taux de succès au FACS
Immunology
22.3K Vues

Test de mort cellulaire : libération du chromium pour mesurer la cytotoxicité
Immunology
151.4K Vues