Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
siRNA Screening to Identify Ubiquitin and Ubiquitin-like System Regulators of Biological Pathways in Cultured Mammalian Cells
W tym Artykule
Podsumowanie
Here, we describe a methodology to perform a targeted siRNA “ubiquitome” screen to identify novel ubiquitin and ubiquitin-like regulators of the HIF1A-mediated cellular response to hypoxia. This can be adapted to any biological pathway where a robust read out of reporter activity is available.
Streszczenie
Post-translational modification of proteins with ubiquitin and ubiquitin-like molecules (UBLs) is emerging as a dynamic cellular signaling network that regulates diverse biological pathways including the hypoxia response, proteostasis, the DNA damage response and transcription. To better understand how UBLs regulate pathways relevant to human disease, we have compiled a human siRNA “ubiquitome” library consisting of 1,186 siRNA duplex pools targeting all known and predicted components of UBL system pathways. This library can be screened against a range of cell lines expressing reporters of diverse biological pathways to determine which UBL components act as positive or negative regulators of the pathway in question. Here, we describe a protocol utilizing this library to identify ubiquitome-regulators of the HIF1A-mediated cellular response to hypoxia using a transcription-based luciferase reporter. An initial assay development stage is performed to establish suitable screening parameters of the cell line before performing the screen in three stages: primary, secondary and tertiary/deconvolution screening. The use of targeted over whole genome siRNA libraries is becoming increasingly popular as it offers the advantage of reporting only on members of the pathway with which the investigators are most interested. Despite inherent limitations of siRNA screening, in particular false-positives caused by siRNA off-target effects, the identification of genuine novel regulators of the pathways in question outweigh these shortcomings, which can be overcome by performing a series of carefully undertaken control experiments.
Wprowadzenie
Modification of proteins with ubiquitin and ubiquitin-like molecules (UBLs) represents an expansive biochemical system that regulates diverse biological pathways and stress responses. The covalent attachment of UBLs to their target proteins can have various outcomes regulating the stability, localization, function or interactome of the substrate1. The enzymatic steps underlying UBL modification were first established for ubiquitin, and now serve as a paradigm for modification with most UBLs, including SUMO, NEDD8, ISG15 and FAT10. For modification to occur, the carboxylate group of the UBL diglycine motif is first activated by an E1 activating enzyme to form a high-energy thiol that is transferred to the active-site cysteine of an E2 conjugating enzyme. The E2 then interacts with a substrate-bound E3 ligase to mediate transfer of the UBL onto (usually) a target lysine residue creating a branched chain (isopeptide) linkage2. Successive rounds of modification can occur to build isopeptide chains onto the substrate, which for ubiquitin can occur through any of its seven lysines, or through its N-terminal methionine to create linear ubiquitin chains. These modifications form discrete topologies with diverse purposes such as creating new interaction motifs and targeting proteins for degradation prior to UBL removal by specialist proteases. In the case of ubiquitin there are two E1 enzymes, 30-40 E2 conjugating enzymes, at least 600 E3 ligases and approximately 100 deubiquitylating enzymes (DUBs). While the pathways are less expansive for the other 10 or so UBLs, the overall ubiquitome complexity affords huge diversity in the biological outcome of a particular UBL modification. However, while major advances in UBL biology have been made, the precise cellular roles of the majority of these ubiquitome components remain unknown.
The use of short interfering ribonucleic acid (siRNAs) has emerged as a powerful tool in reverse genetics due to the ability of siRNAs to specifically target cellular mRNAs for destruction, allowing the role of individual genes to be examined in different biological contexts3. Whole genome screens have been used to identify and validate new regulators of many cellular processes, and have created a wealth of useful data accessible to the wider scientific community. However, while whole genome screens have proven extremely useful, targeted screens are becoming increasingly popular as they are cheaper, faster, involve less data management and report only on members of the genome in which the investigator is most interested. Therefore, to better understand which cellular processes UBL family components are involved in, we have compiled a human siRNA library targeting all known and predicted components of the ubiquitome. This includes the UBLs, E1 activating enzymes, E2 conjugating enzymes, E3 ligases, ubiquitin-binding domain (UBD)-containing proteins and DUBs. This library can be used to screen against a wide range of reporter cell lines of distinct biological problems, thus allowing the unbiased identification of novel UBL components governing these pathways.
The following protocol describes how to perform a rigorous targeted siRNA ubiquitome screen to identify novel regulators of the HIF1A-dependent response to hypoxia. Under normal oxygen tension, HIF1A is subject to prolyl hydroxylation that causes it to be recognized and targeted for degradation by the Von Hippel Lindau (VHL) E3 ligase complex4. Hypoxia inhibits prolyl hydroxylation leading to the stabilization of HIF1A and its subsequent binding to hypoxia response elements (HREs) to drive gene expression. Here, we describe a screen using U20S osteosarcoma cells stably expressing firefly luciferase under the control of three tandem copies of the Hypoxia Response Element (U20S-HRE cells)5. This protocol can be adapted for any biological pathway if a robust read-out of reporter activity is achievable and can be coupled with appropriate positive and negative controls.
Protokół
1. Assay Development Stage
Note: prior to initiating the siRNA screen, an assay development stage is critical to set out important parameters for screening with the reporter cell line. It is essential to invest significant effort at this stage as this will underpin the future success of the screen.
- To characterize the hypoxia-responsiveness of U20S-HRE reporter cell line, grow 2 x 75 cm2 flasks of U20S-HRE cells to 80-90% confluence in Dulbeccos Modified Eagle Medium (DMEM) supplemented with 10% FBS.
- Aspirate the media from one flask of cells, wash twice with 10 ml PBS and detach cells by adding 2 ml 0.05% trypsin-EDTA solution and incubating at 37 °C for 5 min. Resuspend cells in 8 ml DMEM 10% FBS and pipette up and down to create a homogenous cell suspension.
- Pipette 20 μl of the suspension to a cell counting chamber, in duplicate, and insert the chamber into an automated cell counter. Click “Display Image” on the cell counter software and ensure the cells are in focus. Click “Count” and make a note of the cell density. Repeat this for the other duplicate and calculate the average cell density. Dilute the cells with DMEM 10% FBS to a concentration of 60,000 cells/ml.
- Using a multichannel pipette, add 100 μl (6,000 cells) of the diluted cells to the same three columns (e.g. A5-H5, A6-H6 and A7-H7) of 5 sterile white-walled 96-well assay plates, and transfer to a humidified 37 °C incubator at 5% CO2 overnight. Note: these plates will be used to determine the hypoxia-dependent luciferase output for a range of hypoxia exposures: 0 hr, 2 hr, 6 hr, 10 hr and 24 hr.
- At the end of the following day, note the time and add the 24 hr hypoxia plate to the hypoxia workstation set to 1% oxygen. The following morning, calculate the time 10 hr, 6 hr and 2 hr before the 24 hr plate is due to be removed from the hypoxia workstation. At these times, add the 10 hr, 6 hr and 2 hr plates to the hypoxia workstation.
- Prepare 30 ml of 2x combined luciferase lysis/assay buffer (50 mM Tris Phosphate pH 7.8; 16 mM MgCl2; 2 mM DTT; 2% Triton-X-100; 30% glycerol; 1 mM ATP; 1% BSA; 0.25 mM luciferin and 8 μM Na4P2O7). Note: add the components in the order listed and allow the BSA at least 30 min to dissolve before the addition of luciferin and Na4P2O7.
- Remove all plates from the hypoxia workstation at the appropriate time. Add 100 μl of 2x luciferase lysis/assay buffer to the appropriate wells of the assay plate and cover with clear film. Shake the plates on a plate shaker for 10 min at 500 rpm to thoroughly lyse the cells.
- Transfer the plate stack to an automated 96 well plate luminometer rack. Under the “Protocols” menu, select “abs 595” and highlight all wells to be read in the on-screen plate map under the “well selection” menu. Click “run” to measure luminescence of each well then calculate the average reading of each plate. Calculate the hypoxia-dependent fold-increase of reporter activity by dividing the average reading of each hypoxia plate by the average reading of the normoxia (0 hr hypoxia) control plate. Note: it is essential to observe a robust hypoxia-dependent luciferase response to be suitable for screening. A five-ten fold increase in reporter activity is considered excellent.
- Create a master control plate that contains four high control (HIF1A siRNA) replicates (wells A1-D1), four low control (FIH1 siRNA) replicates (wells A12-D12), four buffer only control replicates (wells E1-H1) and four non-target siRNA control replicates (E12-H12), where all siRNA pools are at a concentration of 200 nM. Note: High and low controls are established based on known pathway regulators that increase and decrease the hypoxia response respectively. Alternatively, assay development using a subset of the library may be used to identify suitable controls.
- Using a multichannel pipette, transfer 10 μl of each control siRNA to a sterile white walled assay plate. Prepare transfection mix of 0.1 μl transfection reagent in 10 μl reduced-serum medium (1:100) per well, and transfer 10 μl of transfection mix to each control well. Pipette up and down briefly to mix, and leave the plate to rest for 20-60 min to allow transfection complex formation.
- Prepare a cell suspension of 75,000 cells/ml with the second flask of U20S-HRE cells. Using a multichannel pipette, add 80 μl (6,000 cells) of the cell suspension to each transfection mix. Transfer the plate to a humidified 37 °C incubator with 5% CO2 for 24 hr, then transfer to a hypoxia workstation for a further 24 hr and perform luciferase assays as described in steps 1.6-1.8.
- Calculate the Z-factor for the high and low controls using the formula Z = 1 – [3 x (standard deviation of high control + standard deviation of low control) / (mean of high control – mean of low control)]. Note: a value of between 0.5-1 indicates an excellent assay and should be strived for.
2. Primary Screen
Note: once these basic conditions from the assay development stage are in place, the primary screen can be performed in triplicate in 96 well-plate format using the following protocol.
- Grow 7 x 75 cm2 flasks of U20S-HRE cells to 80-90% confluence in DMEM supplemented with 10% v/v fetal bovine serum (FCS).
Note: the following steps 2.2-2.13 should be carried out on Day 1. - Initiate replicate 1 by preparing a master control plate containing 200 μl of each control as described in step 1.9 and thawing out a dilution series of the siRNA ubiquitome library for a minimum of 30 min. Note: each well contains a pool of 4 siRNAs assembled in 17 x 96-well plates with columns 1 and 12 left empty for controls.
- In a laminar flow hood, label (or bar-code) 17 sterile white walled assay plates with lids from numbers 1-17, corresponding to each plate of the siRNA library series.
- Centrifuge the master control plate and the siRNA ubiquitome library briefly (1 min at 2,000 x g) to ensure all siRNAs accumulate at the bottom of the well.
- Use an automated liquid dispenser to robotically “stamp” 10 μl of each control from the master control plate onto the 17 assay plates, using the same 96-well tip stack for each plate.
- Using a fresh 96-well tip stack for each of the 17 library plates, transfer 10 μl of each ubiquitome siRNA to its corresponding assay plate using an automated liquid dispenser.
- Prepare 40 ml transfection reagent for the 17 assay plates using the ratio established in 1.10. Note: this includes an additional 20 ml to account for the “dead volume” in the cell dispenser. The “dead volume” refers to the volume of liquid continually present within the tubing network in the cell dispenser.
- Use an automated cell dispenser to transfer 10 μl of transfection reagent to each well in the 17 assay plates. Briefly shake the assay plates on a shaker (1 min at 500 rpm) to allow thorough mixing of siRNA and transfection reagent, then leave still at room temperature for 20-60 min to allow siRNA:transfection reagent complex formation.
- Wash two 75 cm2 flasks of U20S-HRE cells twice with 10 ml PBS and detach with 2 ml trypsin-EDTA at 37 °C for 5 min. Add 8 ml DMEM 10% FBS to each flask and pipette up and down several times to create a homogenous cell suspension. Combine cells from both flasks and transfer to a sterile 50 ml plastic tube.
- Calculate cell concentration by pipetting 20 μl cells in duplicate to a cell counting chamber and calculate the average cell concentration from two readings of an automated cell counter as described in step 1.3.
- Prepare 155 ml of cell suspension by diluting with DMEM 10% FBS to a concentration of 75,000 cells per ml in a sterile plastic container. Note: this volume is sufficient for 17 plates and includes an additional 25 ml for the dead volume in the cell dispenser.
- Add a sterile magnetic stirrer to the cell suspension and place on a stirrer set up inside the laminar flow hood to limit cell clumping. Equilibrate the automated cell dispenser with the cell suspension and set it to dispense 80 μl of cells per well (6,000 cells).
- Place each of the 17 plates, in turn, onto the automated cell dispenser and dispense cells. Record the time and stack plates in groups of 5 then place stacks in a humid 37 °C incubator at 5% CO2 for 24 hr. Note: the final concentration of siRNA pool will be 20 nM in 100 μl total volume.
Note: the following steps 2.14-2.15 should be carried out on Day 2. - Initiate the second replicate on day 2, following the procedure outlined in Day 1 for replicate 1 (2.2-2.13).
- After 24 hr of transfection, transfer plates from replicate 1 in a sterile environment to a hypoxia workstation set at 1% oxygen and leave for 24 hr to induce the HRE reporter.
Note: the following steps 2.16-2.20 should be carried out on Day 3. - Initiate the third replicate, following the procedure outlined in Day 1 for replicate 1 (2.2-2.13).
- After 24 hr of transfection, transfer plates from replicate 2 in a sterile environment to a hypoxia workstation set at 1% oxygen and leave for 24 hr to induce the HRE reporter.
- Two hours before replicate 1 plates are to be removed from the hypoxia workstation, prepare 200 ml of 2x luciferase lysis/assay buffer as described in 1.6. Note: this volume includes an additional 20 ml to ensure the buffer reservoir remains well covered.
- Remove replicate 1 assay plates from the hypoxia workstation after 24 hr hypoxia exposure and using an automated liquid dispenser, add 100 μl of 2x luciferase lysis/assay buffer to each assay plate and cover with clear film.
- Shake the plates on a cell shaker for 10 min at 500 rpm to thoroughly lyse the cells. Transfer each plate in turn to a luminometer plate reader and record luminescence as described in step 1.8.
Note: the following steps 2.21-2.22 should be carried out on Day 4. - After 24 hr of transfection, transfer plates from replicate 3 in a sterile environment to a hypoxia workstation set at 1% oxygen and leave for 24 hr to induce the HRE reporter.
- Read the replicate 2 plates by following steps 2.18-2.20 used for replicate 1.
Note: the following step 2.23 should be carried out on Day 5. - Read the replicate 3 plates by following step 2.18-2.20 used for replicate 1.
- Compile all 3 replicates of the primary screen, and calculate the Z factor for each plate using the formula given in 1.12. Note: if any plate has a Z factor of less than 0.5, consider repeating this plate to improve the data quality.
- Calculate the coefficient of variance (CV) for the high and the low controls by using the following formula: 100 x standard deviation of control/ average of control. Note: CV should be as low as possible, it is reasonable to expect 10-15% variation. If variation is significantly higher, consider repeating the plate.
- Calculate percent activation of each siRNA per plate by using the following formula: [(mean of high control – siRNA score) / (mean of high control – mean of low control)] x 100. In addition, calculate the non-target (NT)-fold of each siRNA by using the formula [Sample data/mean of NT]. For both percent activation and NT-fold, calculate the average of the three replicates and compile the values.
- Using the raw data, calculate the Spearman’s Rank Correlation Coefficient (SRCC) to determine how closely the three replicate plates correlate using the formula:
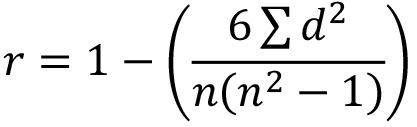
where r = SRCC; d = difference between the two numbers in each pair of ranks and n = number of pairs of data. Note: a good correlation between plate replicates will give values close to 1. If one replicate shows low SRCC against the other 2 plates, investigate further and consider repeating that plate. - Decide which siRNAs are of sufficient interest for secondary screening (up to 80). Employ user-defined cut offs (e.g. selecting all siRNAs showing less than 5% or over 95% percent activation, or less than 0.5 NT-fold or over 1.5 NT-fold) or apply bioinformatics or knowledge-based reasoning to search for clusters of hits falling within the same UBL pathway. Note: normally, a combination of these approaches determines which siRNA are taken into secondary screening.
3. Secondary Screen
Note: a confirmatory secondary screen is carried out based on a maximum of 80 siRNAs of interest from the primary screen. This number can be conveniently carried out with each replicate plated on a single 96 well plate with full controls as per the primary screen (see 2.2). It is very useful to confirm that the regulators identified in the primary screen reproducibly elicit the same phenotype, and this will assist in refining the triage decisions on which hits should be carried through to the final tertiary/deconvolution screen.
- Grow 1 x 75 cm2 flask of U20S-HRE cells to 80-90% confluence in DMEM with 10% FBS.
- Note the plate number and location of the primary screen hits, and plan their new location on the secondary screen cherry picked master plate, leaving columns 1 and 12 free for controls. Prepare the control master plate as in 1.9 but with a total volume of each control of 50 μl.
- Thaw a second dilution series of the ubiquitome library (set aside for cherry-picking individual siRNA pools) for a minimum of 30 min. Centrifuge the plates briefly (1 min at 2,000 x g) then add 50 μl of the secondary screen siRNAs to their new plate locations on the cherry picked master plate.
- Carry out the secondary screen using the same basic protocol outlined for the primary screen with the following modifications: (a) run all three replicates on the same day in one assay plate per replicate and (b) adjust volumes of reagents accordingly.
4. Tertiary/Deconvolution Screen
Note: tertiary or deconvolution screening is performed on a maximum of 20 siRNAs from the secondary screen. This step is to examine the effect of knockdown using each individual siRNA from the original pool of four. Normally, at least two individual siRNA duplexes from each pool should illicit the same phenotype to have a reasonable degree of confidence that the observed phenotype is not due to siRNA off-target effects. To further increase confidence at this stage, additional individual siRNA duplexes targeting the gene in question can be designed and tested for their ability to elicit the given phenotype. Results from these experiments may then be used to calculate the H score, where H = 0.6 or over (i.e. where at least 3 out of 5 individual siRNAs elicit the phenotype) is considered acceptable6.
- Grow one 75 cm2 flask of U20S-HRE cells to 80-90% confluence in DMEM with 10% FBS.
- Create a plate map for the tertiary screen by planning the location of the four individual siRNA duplexes of each hit on the tertiary screen master plate, leaving columns 1 and 12 free for controls. Prepare the control master plate as in 1.9 but with a total volume of each control of 50 μl.
- Thaw the deconvolution/individual siRNA plates and add 50 μl of the individual siRNAs (200 nM) into each well of the tertiary screen master plate according to the plate map (4.2) to allow for three x 10 μl replicates and a comfortable 20 μl excess.
- Carry out the tertiary screen using the same basic protocol outlined for the primary screen with the following modifications: (a) run all three replicates on the same day in one assay plate per replicate and (b) adjust volumes of reagents accordingly
Note: Ideally the threshold for individual siRNA duplexes should be set at the same stringency cut-off as for the pool. However, it may be acceptable to relax the threshold by 10-20% for individual siRNAs, especially where at least one other duplex falls within the set threshold. It is worthwhile bearing in mind that the individual effect of siRNAs may be less than that of the pool, and conversely, in some cases individual siRNAs may show a stronger effect on the cellular phenotype in isolation than when it exists in the pool (even when concentration is accounted for).
Wyniki
Prior to screening, the hypoxia-responsiveness of U20S-HRE cells is established. U20S-HRE cells express a reporter construct consisting of firefly luciferase fused downstream of three tandem copies of the hypoxia response element, which is bound by the HIF1A/HIF1B heterodimer upon exposure to hypoxia (Figure 1A). Cells are placed in a hypoxia workstation for a range of times to establish which hypoxia exposure produces the most effective response for screening. U20S cells express low le...
Dyskusje
The use of genome-wide siRNA screens in mammalian cells has proven to be extremely valuable in identifying novel regulators of distinct biological pathways. Here, we have described the use of a targeted ubiquitome siRNA screen to identify regulators of the HIF1A-mediated cellular response to hypoxia. Targeted screens are becoming increasingly attractive as they are generally cheaper, quicker, easier to manage and report only on the pathway components in which the investigators are interested 7, 10, 11.
Ujawnienia
The authors have nothing to disclose.
Podziękowania
This work was supported by The Wellcome Trust, Glaxosmithkline (GSK) and the Scottish Institute for Cell Signalling (now part of the MRC Protein Phosphorylation and Ubiquitylation unit).
Materiały
| Name | Company | Catalog Number | Comments |
| Automated Liquid Dispenser | Fluid-X | XPP-721 | http://www.fluidx.eu/BIOTRACK/xpp-721-liquid-handling-system.html |
| White Walled Assay Plate | Greiner Bio One | 655083 | http://www.greinerbioone.com/en/row/articles/catalogue/article/37_11/13221/ |
| Clear Plate Film | Perkin Elmer | 1450-461 | http://www.perkinelmer.co.uk/Catalog/Product/ID/1450-461 |
| siRNA library | Thermo Scientific | On-Target Plus | http://www.thermoscientificbio.com/rnai-and-custom-rna-synthesis/sirna/on-targetplus-sirna/search-gene/ |
| Transfection reagent | Invitrogen | Lipofectamine RNAimax | http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Protein-Expression-and-Analysis/Transfection-Selection/lipofectamine-rnaimx.html |
| Reduced Serum Medium | Invitrogen | Optimem | http://products.invitrogen.com/ivgn/product/31985062?ICID=search-product |
| DMEM | Invitrogen | 41965-039 | http://products.invitrogen.com/ivgn/product/41965039# |
| FBS | Invitrogen | 16000-044 | https://products.invitrogen.com/ivgn/product/16000044?ICID=search-product# |
| Tryspin-EDTA | Invitrogen | 25300-054 | https://products.invitrogen.com/ivgn/product/25300054?ICID=search-product# |
Odniesienia
- Hochstrasser, M. Origin and function of ubiquitin-like proteins. Nature. 458, 422-429 (2009).
- Schulman, B. A., Harper, J. W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 10, 319-331 (2009).
- Siomi, H., Siomi, M. C. On the road to reading the RNA-interference code. Nature. 457, 396-404 (2009).
- Kaelin, W. G., Ratcliffe, P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 30, 393-402 (2008).
- Melvin, A., Mudie, S., Rocha, S. The chromatin remodeler ISWI regulates the cellular response to hypoxia: role of FIH. Mol Biol Cell. 22, 4171-4181 (2011).
- Bhinder, B., Djaballah, H. A simple method for analyzing actives in random RNAi screens: introducing the "H Score" for hit nomination & gene prioritization. Comb Chem High Throughput Screen. 15, 686-704 (2012).
- Bett, J. S., et al. The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability. Biochem J. 451, 185-194 (2013).
- Zhang, J. H., Chung, T. D., Oldenburg, K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 4, 67-73 (1999).
- Birmingham, A., et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 6, 569-575 (2009).
- Stagg, H. R., et al. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol. 186, 685-692 (2009).
- Zhang, Y., et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 13, 623-629 (2011).
- Jackson, A. L., et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 21, 635-637 (2003).
- Birmingham, A., et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 3, 199-204 (2006).
- . Whither RNAi. Nat Cell Biol. 5, 489-490 (2003).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone