Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
The Terroir Concept Interpreted through Grape Berry Metabolomics and Transcriptomics
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
This article describes the application of untargeted metabolomics, transcriptomics and multivariate statistical analysis to grape berry transcripts and metabolites in order to gain insight into the terroir concept, i.e., the impact of the environment on berry quality traits.
Streszczenie
Terroir refers to the combination of environmental factors that affect the characteristics of crops such as grapevine (Vitis vinifera) according to particular habitats and management practices. This article shows how certain terroir signatures can be detected in the berry metabolome and transcriptome of the grapevine cultivar Corvina using multivariate statistical analysis. The method first requires an appropriate sampling plan. In this case study, a specific clone of the Corvina cultivar was selected to minimize genetic differences, and samples were collected from seven vineyards representing three different macro-zones during three different growing seasons. An untargeted LC-MS metabolomics approach is recommended due to its high sensitivity, accompanied by efficient data processing using MZmine software and a metabolite identification strategy based on fragmentation tree analysis. Comprehensive transcriptome analysis can be achieved using microarrays containing probes covering ~99% of all predicted grapevine genes, allowing the simultaneous analysis of all differentially expressed genes in the context of different terroirs. Finally, multivariate data analysis based on projection methods can be used to overcome the strong vintage-specific effect, allowing the metabolomics and transcriptomics data to be integrated and analyzed in detail to identify informative correlations.
Wprowadzenie
Large-scale data analysis based on the genomes, transcriptomes, proteomes and metabolomes of plants provides unprecedented insight into the behavior of complex systems, such as the terroir characteristics of wine which reflect the interactions between grapevine plants and their environment. Because the terroir of a wine can be distinct even when identical grapevine clones are grown in different vineyards, genomics analysis is of little use because the clonal genomes are identical. Instead it is necessary to look at correlations between gene expression and the metabolic properties of the berries, which determine the quality traits of wine. The analysis of gene expression at the level of the transcriptome benefits from the similar chemical properties of all transcripts, which facilitates quantitative analysis by exploiting universal characteristics such as hybridization to immobilized probes on microarrays. In contrast, universal analytical methods in proteomics and metabolomics are more challenging because of the huge physical and chemical diversity of individual proteins and metabolites. In the case of metabolomics this diversity is even more extreme because individual metabolites differ vastly in size, polarity, abundance and volatility, so no single extraction process or analytical method offers a holistic approach.
Among the analytical platforms suitable for non-volatile metabolites, those based on high performance liquid chromatography coupled to mass spectrometry (HPLC-MS) are much more sensitive than alternatives such as HPLC with ultraviolet or diode array detectors (HPLC-UV, HPLC-DAD) or nuclear magnetic resonance (NMR) spectroscopy, but quantitative analysis by HPLC-MS can be influenced by phenomena such as the matrix effect and ion suppression/enhancement1-3. The investigation of such effects during the analysis of Corvina grape berries by HPLC-MS using an electrospray ionization source (HPLC-ESI-MS), showed that sugars and other molecules with the lowest retention times were strongly underreported, probably also reflecting the large number of molecules in this zone, and that the abundance of other molecules could be underestimated, overestimated or unaffected by the matrix effect, but the data normalization for the matrix effect seemed to have limited impact on the overall results4,5. The method described herein is optimized for the analysis of medium-polarity metabolites that accumulate at high levels in grape berries during ripening, and which are significantly impacted by the terroir. They include anthocyanins, flavonols, flavan-3-ols, procyanidins, other flavonoids, resveratrol, stilbenes, hydroxycinnamic acids and hydroxybenzoic acids, which together determine the color, taste and health-related properties of wines. Other metabolites, such as sugars and aliphatic organic acids, are ignored because quantitation by HPLC-MS is unreliable due to the matrix effect and ion suppression phenomena5. Within the polarity range selected by this method, the approach is untargeted in that it aims to detect as many different metabolites as possible6.
Transcriptomics methods that allow thousands of grapevine transcripts to be monitored simultaneously are facilitated by the availability of the complete grapevine genome sequence7,8. Early transcriptomics methods based on high-throughput cDNA sequencing have evolved with the advent of next-generation sequencing into a collection of procedures collectively described as RNA-Seq technology. This approach is rapidly becoming the method of choice for transcriptomics studies. However, a large body of literature based on microarray, which allow thousands of transcripts to be quantified in parallel by hybridization, has accumulated for grapevine. Indeed, before RNA-Seq became a mainstream technology, many dedicated commercial microarray platforms had been developed allowing grapevine transcriptome to be inspected in great detail. Among the vast variety of platforms, only two allowed genome-wide transcriptome analysis9. The most evolved array allowed the hybridization of up to 12 independent samples on a single device, thus reducing the costs of each experiment. The 12 sub-arrays each comprised 135,000 60-mer probes representing 29,549 grapevine transcripts. This device has been used in a large number of studies10-24. These two platforms have now been discontinued but a new custom microarray has recently been designed and represents a more recent development as it contains an even greater number of probes representing additional newly discovered grapevine genes25.
The large-sale datasets produced by transcriptomics and metabolomics analysis require suitable statistical methods for data analysis, including multivariate techniques to determine correlations between different forms of data. The most widely used multivariate techniques are those based on projection, and these can be unsupervised, such as principal component analysis (PCA), or supervised, such as bidirectional orthogonal projection to latent structures discriminant analysis (O2PLS-DA)26. The protocol presented in this article utilizes PCA for exploratory data analysis and O2PLS-DA to identify differences between groups of samples.
Protokół
1. Select Appropriate Materials and Construct a Sampling Plan
- Begin the experiment by developing an appropriate sampling plan. There is no generic and universal approach so evaluate each plan on a case-by-case basis. Ensure that the sampling plan states the sampling places, times and the precise sampling procedure. See Figure 1 for the sampling plan used in this case study.
NOTE: In this case study, grape berries from a single clone (Vitis vinifera cv. Corvina, clone 48) were collected from seven commercial vineyards in three different macro-zones in the province of Verona (Lake Garda, Valpolicella and Soave). The principal features of each vineyard are summarized in Table 1. Berries were collected during three growing seasons (2006, 2007, and 2008) at three time points, corresponding to veraison (the onset of ripening), mid-ripening and ripe berries.- For each of the accessions (vineyard/year/ripening stage), harvest 30 clusters from different positions along two vine rows, with randomized heights and locations on the plant.
- Select three berries randomly from each cluster, avoiding those with visible damage and/or signs of infection.
- Repeat steps 1.1.1 and 1.1.2 to obtain three independent pools.
- Deseed the berries and freeze the pericarp immediately in liquid nitrogen.
- Crush 10 frozen berries from each pool with an automatic mill grinder, and divide each powdered sample into two equal parts, one for transcriptomics analysis and one for metabolomics analysis.
- Store the powders at -80 °C.
| AM | BA | BM | CS | FA | MN | PM | |
| Macro-zone | Soave | Lake Garda | Valpolicella | Lake Garda | Valpolicella | Valpolicella | Soave |
| Height (m) | 250 | 120 | 450 | 100 | 130 | 250 | 130 |
| Rootstock | 41B | S04 | K5BB | 420A | 420A | K5BB | 41B |
| Row direction | E-W | N-S | E-W | E-W | E-W | N-S | N-S |
| Training system | Overhead System (Pergola) | Overhead System (Pergola) | Vertical Shoot Positioning (Guyot) | Overhead System (Pergola) | Overhead System (Pergola) | Vertical Shoot Positioning (Guyot) | Vertical Shoot Positioning (Guyot) |
| Soil type | Silty clay | Loam | Clay | Loam | Clay Loam | Silt loam | Clay loam |
| Planting layout (m) | 3.20 x 1.00 | 4.50 x 0.80 | 4.00 x 1.25 | 3.50 x 1.20 | 3.50 x 0.75 | 2.80 x 1.00 | 1.80 x 0.80 |
| Total lime % | 3.9 | 19.3 | 18.3 | 14.4 | 31 | 5.9 | 27.9 |
| Active lime % | 0.5 | 2.6 | 9.4 | 6.3 | 11.3 | 3.1 | 8.3 |
| Sand % | 15 | 47 | 66 | 42 | 29 | 13 | 36 |
| Loam % | 43 | 36 | 21 | 37 | 39 | 67 | 36 |
| Clay % | 42 | 17 | 13 | 21 | 32 | 20 | 28 |
| Soil pH | 8.3 | 7.9 | 7.8 | 8.2 | 8.2 | 7.8 | 7.9 |
| Organic substance (%) | 2.9 | 2.5 | 2.2 | 1.2 | 2.9 | 1.6 | 2.5 |
| Exchangeable phosphorus (mg/kg) | 26 | 73 | 73 | 68 | 48 | 47 | 64 |
| Exchangeable potassium (mg/kg) | 190 | 376 | 620 | 230 | 168 | 154 | 126 |
| Exchangeable magnesium (mg/kg) | 272 | 468 | 848 | 623 | 294 | 293 | 183 |
| Exchangeable calcium (mg/kg) | 6500 | 5380 | 7358 | 6346 | 4652 | 10055 | 2878 |
| Berry Reducing Sugars 2006 | 211.25 ± 1.20 | 176.20 ± 0.42 | 187.40 ± 0.00 | 203.70 ± 1.13 | 212.55 ± 0.64 | 195.20 ± 0.00 | 211.65 ± 0.64 |
| Berry Reducing Sugars 2007 | 190.00 ± 1.27 | 165.25 ± 0.49 | 153.00 ± 0.42 | 203.60 ± 0.71 | 210.90 ± 0.71 | 192.25 ± 0.64 | 188.70 ± 1.84 |
| Berry Reducing Sugars 2008 | 191.35 ± 0.64 | 178.90 ± 0.57 | 170.05 ± 0.49 | 205.15 ± 1.48 | 188.70 ± 0.57 | 169.35± 0.49 | 108.05 ± 1.06 |
| Berry pH 2006 | 3.01 ± 0.01 | 2.96 ± 0.01 | 2.84 ± 0.00 | 2.9 ± 0.00 | 2.98 ± 0.00 | 3.02 ± 0.00 | 3.06 ± 0.01 |
| Berry pH 2007 | 2.97 ± 0.00 | 3.00 ± 0.00 | 2.74 ± 0.00 | 3.07 ± 0.01 | 2.98 ± 0.00 | 2.87 ± 0.01 | 3.09 ± 0.00 |
| Berry pH 2008 | 2.83 ± 0.00 | 3.04 ± 0.01 | 2.71 ± 0.00 | 2.98 ± 0.01 | 2.98 ± 0.00 | 2.82 ± 0.00 | 3.11 ± 0.00 |
| Harvesting Date | 2006 | 2007 | 2008 | ||||
| Veraison | 8-Aug | 18-Jul | 12-Aug | ||||
| Mid Ripening | 4-Sep | 8-Aug | 2-Sep | ||||
| Ripe | 18-Sep | 29-Aug | 23-Sep |
Table 1: Principal features of each vineyard and sample collection dates. m = meters, E-W = Eat-West, N-S = North-South.
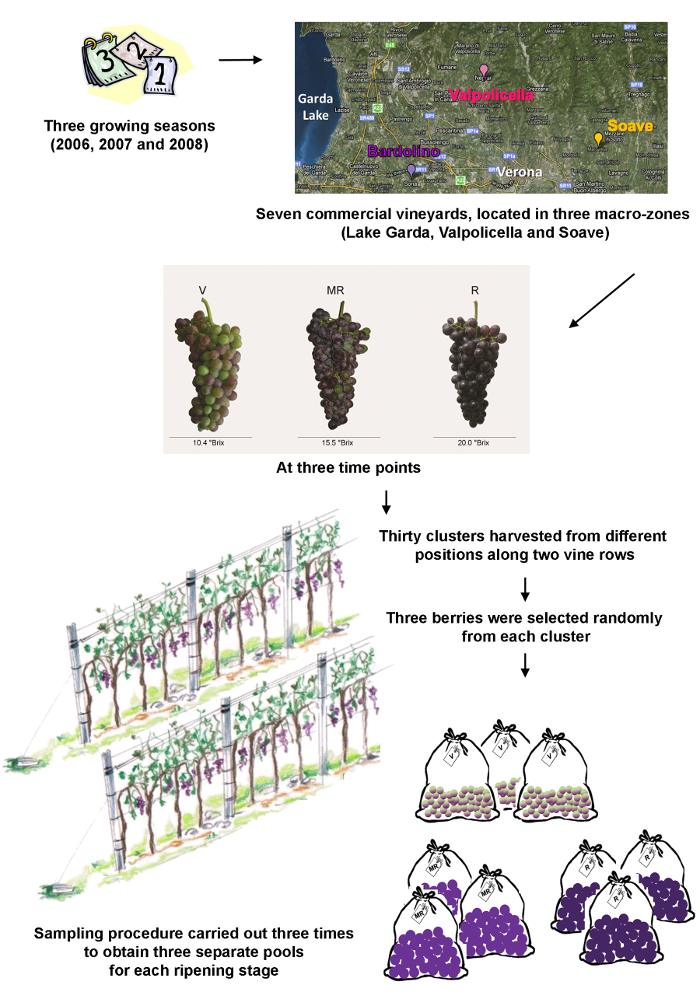
Figure 1: Schematic representation of the sampling procedure. The three wine production macro-zones are located in the surroundings of the city of Verona, Veneto region, Italy. The three time points are veraison (V) representing the onset of ripening in viticulture, mid-ripening (MR) and ripe berries (R). Please click here to view a larger version of this figure.
2. Prepare Berry Powder Extracts, Analyze the Metabolites and Process the Data
- Prepare the Berry Powder Samples for Analysis.
- Prepare the metabolic extracts of the berry samples at room temperature in three volumes (w/v) of methanol acidified with 0.1% (v/v) formic acid in an ultrasonic bath at 40 kHz for 15 min. Use LC-MS-grade water and formic acid, and HPLC-grade methanol.
- Centrifuge the extracts at 16,000 x g for 10 min at 4 °C.
- Dilute the supernatant from step 2.1.2 in two volumes (v/v) of deionized water and pass through a 0.2-µm filter.
- Analyze the Berry Extracts by HPLC-ESI-MS.
- Set up the HPLC-ESI-MS system according to the supplier's recommendations.
NOTE: In this case study, the setup comprised an HPLC system equipped with an autosampler, connected in-line with an ion trap mass spectrometer and electrospray ionization source. - Connect the HPLC system with a C18 guard column (7.5 x 2.1 mm2) and a Reversed Phase C18 column (150 x 2.1 mm2, particle size 3 µm).
- Prepare the solvents used as the mobile phase. Use 5% (v/v) acetonitrile, 0.5% (v/v) formic acid in water as Solvent A and 100% acetonitrile as solvent B. Use LC-MS-grade acetonitrile, water and formic acid.
- Run the HPLC separation with a linear gradient of solvents A and B at a constant flow rate of 0.2 ml min-1 using a sample injection volume of 30 µl.
- Initially equilibrate the column with 100% solvent A. Following sample injection, establish a gradient from 0% to 10% solvent B in 5 min, from 10% to 20% solvent B in 20 min, from 20% to 25% solvent B in 5 min, and from 25% to 70% solvent B in 15 min.
- Analyze each sample in duplicate. Randomize sample analysis to avoid instrument-driven effects. Allow 20 min for re-equilibration with 100% solvent A between each analysis.
- Acquire mass spectra in alternate negative and positive ionization modes. For the results described in this case study, set the parameters as in Table 2. The precise machine setting depends on the specific platform.
NOTE: Alternatively, use other suitable methods according to the specific platform.- In all cases, as with any separation platform, make sure to use a suitable re-equilibration time, in order to have retention time reproducibility. When the number of samples is high (above ten samples), analyze them in batches of 9-10 samples, with chromatographic column cleaning programs (slow gradients between the two elution solvents) between each batch. To have enough retention time reproducibility, ensure that the first chromatographic analysis of each batch is a blank analysis (i.e., analysis of the solvent).
- Also acquire mass spectra in fragmented negative and positive ion modes setting the fragmentation options (two ion precursors, MS3). Annotate the metabolites by fragmentation tree analysis (MS/MS and MS3) according to manufacturer's instructions.
NOTE: This is particularly suitable for plant metabolites because they are often glycosylated, therefore the first fragmentation (MS/MS) tends to remove the sugar moieties leaving the free aglycone ion, and subsequent fragmentation (MS3) helps to identify the aglycone against an authentic standards library. - Acquire MS/MS and MS3 spectra in the m/z range 50-1,500 with a fragmentation amplitude of 1 V. Alternatively, use other suitable methods according to the specific platform.
- For each m/z signal, compare the MS/MS and MS3 fragmentation patterns and retention times to the authentic standards library according to manufacturer's instructions.
NOTE: Many type of platforms include software facilities to build this kind of library through the analysis of authentic standards. This should identify many of the signals but not all signals will be annotated. - For the remaining anonymous signals, compare the MS/MS and MS3 fragmentation patterns with those published in the literature, searching for m/z values (i.e., digit "m/z 353" to look for publications in which molecules with such m/z are mentioned) with any of the common search engine freely available, or use online databases such as MassBank (www.massbank.jp/en/database.html) and the Human Metabolome Database (www.hmdb.ca/search/spectra?type=ms_search).
- Set up the HPLC-ESI-MS system according to the supplier's recommendations.
| Mass spectrometer components | Function | Parameters |
| Electrospray Ionization Source | Nebulizing Gas | 50 psi, 350 °C |
| Drying gas | 10 L min-1 | |
| Ion trap and detector | Scan | Full scan mode, 13,000 m/z per second, range 50-1,500 m/z |
| Target mass | 400 m/z | |
| Collision Gas | Helium | |
| Vacuum pressure | 1.4 x 10-5 mbar | |
| Capillary source | +4,000 V | |
| End plate offset | -500 V | |
| Skimmer | -40 V | |
| Cap exit | -121 V | |
| Oct 1 DC | -12 V | |
| Oct 2 DC | -1.7 V | |
| Lens 1 | +5 V | |
| Lens 2 | +60 V | |
| ICC for positive ionization mode | 20.000 | |
| ICC for negative ionization mode | 7,000 |
Table 2: Principal parameter set for acquiring mass spectra.
| Operation | Selection | Function | Parameters | Values |
| Peak Detection | Mass detection | Centroid | Noise level | 3,500 |
| Chromatogram builder | Highest data point | min time span | 0.15 | |
| min height | 4,000 | |||
| m/z tolerance | 0.3 | |||
| Peak Detection | Peak deconvolution | Local minimum search | Chromatographic threshold | 70 |
| Search minimum in RT range (min) | 0:50 | |||
| Minimum relative height | 15% | |||
| Minimum absolute height | 4,000 | |||
| Min ratio of peak top/edge | 2 | |||
| Duration range (min) | 0-10 | |||
| Isotopes | Isotopic peaks grouper | - | m/z tolerance | 1.2 |
| RT tolerance | 0:50 | |||
| monotonic shape | No | |||
| Maximum charge | 3 | |||
| Representative isotope | No | |||
| Alignment | Join aligner | - | m/z tolerance | 1.2 |
| Weight for m/z | 10 | |||
| Retention time tolerance | 0:50 | |||
| Weight for RT | 5 | |||
| Require same charge state | No | |||
| Require same ID | No | |||
| Compare isotope pattern | No | |||
| Gap filling | Peak finder | - | Intensity tolerance | 20% |
| m/z tolerance | 0.9 | |||
| Retention time tolerance | 0:40 | |||
| RT correction | No | |||
| Filtering | Duplicate Peak filter | - | m/z tolerance | 1.2 |
| RT tolerance | 0:30 | |||
| Require same identification | No |
Table 3: Mzmine workflow with specific values to process negative LC-MS grape berry data files.
- Process the LC-MS Data.
- Access or download a data processing software package that can extract relevant information from many raw chromatograms and build a data matrix in which each detected metabolite is quantified in each sample.
NOTE: The protocol steps below are tailored for the open-source software MZmine v2.14 (http://mzmine.sourceforge.net). It is an open-source software for mass-spectrometry data processing, with the main focus on LC-MS data.27 - Transform the LC-MS chromatogram data into netCDF format using the software provided by the equipment manufacturer. For the results described here, use the Bruker Daltonics Esquire v5.2 and Data Analysis v3.2 softwares and perform steps according to the manufacturer's instructions.
NOTE: Other converters may be used (various free-ware converters are available). - Import the .cdf files into the software.
- Implement the peak detection, alignment, gap filling and peak filtering procedures with the parameters reported in Table 3.
- As first step, select an imported .cdf file, then go to Visualization → TIC/XIC Visualizer. Put the cursor upon the base of the smallest peak in the chromatogram and take note about the minimum intensity signal of the base ion. Then go to Raw Data Methods → Peak Detection.
- Select Mass Detection and fill the signal level to detect individual ions for each scan and create an ion list.
NOTE: Check the algorithm prior to performing the mass detection - it depends on the mass spectrometer. In our case, we select Centroid. - Select Chromatogram Builder and fill with the signal level to connect data points from the ion list and build a chromatogram for each mass value. See the parameters specified in the mass spectrometer's manual to adjust the Min Time Span and m/z tolerance.
- Next, go to Peak List Methods → Peak Detection → Chromatogram Deconvolution. Select the appropriate algorithm (in this case Local Minimum Search) to allow the separation of each chromatogram into individual peaks.
- Manually inspect the chromatograms to find out the appropriate values for the following parameters. Set the Chromatographic Threshold to 30% to remove the noise and Search Minimum in RT Range to 2 (min) to identify the presence of minimum locals to discriminate two peaks.
- Set Minimum Relative Height at 2.0. Manually inspect the chromatogram to identify the minimum absolute height of signal that corresponds to a peak and not to the background; set this value as Absolute Height (for instance, 10,000 in the presented experiment).
- Set Min Ratio of Peak Top/Edge to 1.1 to state the minimum ratio between the top and the lowest data point intensities of a peak to be recognized as a true peak.
- Manually inspect the chromatograms to see which is the minimum duration of the various peaks in the used chromatographic conditions (for instance, between 0.2 and 2 min, depending on the compound, in the presented experiments), and consequently use these values as Peak Duration Range (min) to set the range of acceptable peak length as duration.
- Then go to Peak List Methods → Isotopes → Isotopic Peaks Grouper to group isotopes in one peak, usually in the most intense. Note: The parameters depend on the resolution of the mass spectrometer and the reproducibility of the retention times.
- Finally, go to Peak List Methods → Alignment → Join Aligner to align peaks depending on their m/z and retention time by using a match score.
- After aligning the peaks, fill any data gaps by clicking on Peak List Methods → Gap Filling → Peak Finder. Finally, filter duplicate data points by clicking on Peak List Methods → Filtering → Duplicate Peak Filter.
- Export the resulting dataset as a .csv file.
- Manually change the .csv extension to a .txt extension. If no file extensions are visible, change the computer settings to reveal file extensions. Go to File Explorer Options → View → Advanced Settings and uncheck the box 'Hide extensions for known file types'.
- Import the .txt files into a spreadsheet. This produces a data matrix in which all the detected metabolites, recognized by an identification number, m/z value and retention time, are quantified in all samples in terms of their peak area values.
- Access or download a data processing software package that can extract relevant information from many raw chromatograms and build a data matrix in which each detected metabolite is quantified in each sample.
3. Prepare Berry Powder Extracts for Transcriptome Analysis and Process the Data
- Extract Total RNA from the Berry Samples and Determine the RNA Quality.
- Extract total RNA from the berry samples.
NOTE: In this case study, a proprietary kit and a modified procedure that ensures the complete removal of molecules that interfere with RNA analysis, such as polysaccharides and polyphenols, were used. The instructions below are specific for this kit18.- Weigh 400 mg of berry powder for each sample, divide it into two portions and place 200 mg each in two microfuge tubes.
- Add 900 µl of the lysis solution containing β-mercaptoethanol (provided in the kit) to each 200 mg of powder and vortex immediately and vigorously for at least 30 sec. Heat the sample at 56 °C for 5 min shaking at 800 rpm.
- Centrifuge the samples at maximum speed in a benchtop microfuge for 10 min to pellet cellular debris.
- Pipet 700 µl of the supernatant into the filtration column provided in the kit (blue retainer ring) seated in a 2 ml collection tube. Close the cap and centrifuge at maximum speed in a benchtop microfuge for 1 min to remove residual debris. Repeat this step twice using the same filtration column but a fresh collection tube, resulting in three tubes each containing ~700 µl of the clarified lysate.
- Pipet 750 µl of binding solution (supplied in the kit) into each tube of clarified lysate and mix immediately by pipetting up and down at least five times. Transfer 700 µl of this mixture to the binding column provided in the kit (red retainer ring) seated in a 2 ml collection tube. Close the cap and centrifuge at maximum speed in a benchtop microfuge for 1 min to bind the RNA.
- Decant the flow-through fraction, invert the collection tube and tap it briefly on a clean pad of absorbent paper to drain the residual liquid.
- Return the column to the collection tube and pipet the remaining mixture into the same column and repeat the centrifugation and decanting steps. Repeat until the entire mixture has been filtered into the same red binding column.
- Now follow the remaining instructions in the kit and elute the RNA in 50 µl elution buffer (supplied in the kit) and store it at -80 °C ready for the quality control steps.
- Determine the quantity and purity of the RNA using a spectrophotometer. Record the absorbance ratios which reveal the degree of contamination with proteins (A260/280) and polyphenols/polysaccharides (A260/230).
NOTE: RNA suitable for microarray hybridization should score at least 1.8 for both ratios. - Determine the integrity of the RNA.
NOTE: Various systems can be used. A digital acquirer that performs a capillary electrophoretic run in combination with a fluorescent dye was used in this case study. RNA suitable for microarray hybridization should have a RNA Integrity Number (RIN) of at least 8.
- Extract total RNA from the berry samples.
- Prepare Samples and Hybridize the RNA to a Custom Microarray.
- Set the starting amount of total RNA for microarray analysis to 200 ng by diluting the RNA solution obtained in the step 3.1.1.8 with deionized RNase-free water. Add the appropriate amount of RNA to a 1.5 ml microcentrifuge tube in a final volume of 1.5 µl.
- Add 2 µl of diluted spike mix to each RNA sample and follow the manufacturer's instructions to synthesize first strand cDNA, transcribe this into cRNA and label the cRNA with cyanine 3CTP.
- Purify the labeled cRNA according to the manufacturer's instructions and elute in 30 µl RNase-free water.
- Determine the yield and specific activity of each cRNA by recording three values on a spectrophotometer: cyanine 3 dye concentration (pmol µl-1), RNA purity (A260/280) and cRNA concentration (ng µl-1). Use the formulae found in the manufacturer's instruction manual to calculate the cRNA yield (µg) and the specific activity (pmol Cy3 per µg cRNA).
NOTE: Recommended yields and specific activities differ on the basis of the specific microarray format. In this case study a 4-pack 44K format was chosen, the recommended yield was 1.65 and the specific activity was 9. - Design the custom microarray using suitable software for probe design.
- For the results described in this case study, prepare a new custom microarray, designed on the 4-pack 44K format by using a web-based application for custom microarray designs and oligonucleotide libraries. Design probes to match 34,651 target transcripts, including 29,971 predicted transcripts from the Pinot noir V1 array, 4,500 new loci identified in the Pinot cultivar by Corvina transcriptome reconstruction and 180 private Corvina genes25.
NOTE: This involved the production of 34,651 specific 60-mer probes, comprising 29,798 Pinot noir predicted transcripts, 4,392 new Pinot loci and 179 Corvina private genes.
- For the results described in this case study, prepare a new custom microarray, designed on the 4-pack 44K format by using a web-based application for custom microarray designs and oligonucleotide libraries. Design probes to match 34,651 target transcripts, including 29,971 predicted transcripts from the Pinot noir V1 array, 4,500 new loci identified in the Pinot cultivar by Corvina transcriptome reconstruction and 180 private Corvina genes25.
- Prepare the hybridization assembly based on 4-pack format specifications as follows.
- Place 1.65 µg of Cy3-labeled cRNA in a final volume of 41.8 µl of deionized RNase-free water. Add 11 µl of 10x Blocking Agent and 2.2 µl 25x Fragmentation Buffer. Incubate at 60°C for 30 min in a thermostatic bath to fragment RNA. Immediately cool on ice for 1min.
- Finally, add 55 µl of 2x hybridization buffer, mix well by pipetting and spin for 1 min at 15,500 x g at RT. Promptly place the microcentrifuge tube on ice. Use immediately, do not store it.
- Load the custom microarray as follows.
- Load a gasket slide with the label facing up into the base of a hybridization chamber. Slowly load 100 µl of hybridization sample obtained in step 3.2.6.2 onto each gasket well, dispensing the liquid with the tip of the pipette, avoiding bubbles.
- Slowly place the custom microarray facing down, ensuring that the numeric barcode is facing up. Make sure that the sandwich pair is properly aligned. Finally place the cover of the hybridization chamber onto the sandwiched slides and hand-tighten the clamp onto the chamber. Rotate the assembled chamber to assess the mobility of bubbles.
- Place assembled slide chamber in a rotisserie in a hybridization oven set to 65 °C. Set the rotator to rotate at 10 rpm. Allow hybridization to proceed for 17 hr.
- Proceed with the microarray slide wash as follows.
- First, prepare three slide-staining dishes and fill them with the appropriate washing buffers: 2 dishes with Wash Buffer 1 at RT and 1 dish with pre-warmed (37 °C) Wash Buffer 2.
- Disassemble the hybridization chamber and remove the sandwich. With the microarray slide numeric barcode facing up, submerge the sandwich into the first slide-staining dish filled with Wash Buffer 1 at RT and, with the help of clean forceps, separate the gasket from the microarray slide. Quickly transfer the microarray slide into a slide rack and place it in the second slide-staining dish filled with Wash Buffer 1 at RT.
- Put the slide-staining dish onto a magnetic stirrer and wash 1 min with moderate stirring. Quickly transfer the slide rack into the third slide-staining dish filled with pre-warmed (37 °C) Wash Buffer 2 and wash 1 min with moderate stirring.
- Slowly remove the rack from the slide-staining dish and carefully remove the slide from the rack, avoiding droplets.
NOTE: Do not add Triton X-102 to the washing buffers and omit the acetonitrile washing step.
- Store the washed chip in the dark at room temperature.
- Scan the Microarray and Extract the Salient Features.
- Place the microarray slide into a suitable scanner and scan each array using the parameter settings recommended in the microarray manufacturer's instructions manual. For the results described here, place each microarray slide into a Slide Holder to facilitate the scanning procedure.
- Import the output .shp file in an appropriate software that is able to convert a digital signal into numeric fluorescent values. Check the quality control report to ensure that the hybridization procedure was successful.
- For the results described here, use the parameter settings recommended in the instruction manual of the feature extraction software and inspect the quality control report to ensure that the parameters listed in Table 4 are within the normal range given by the manufacturer.
| Metric Name | Upper Limit | Lower Limit | Description |
| AnyColorPrcntFeatNonUnif | 1.00 | NA | Percentage of features that are feature non-uniformity outliers in either channel |
| DetectionLimit | 2.00 | 0.10 | Average plus 1 standard deviation of the spike ins below the linear concentration range |
| absGE1E1aSlope | 1.20 | 0.90 | Absolute of slope of fit for Signal vs. Concentration of E1a probes |
| MedCVProcSignal | 8.00 | NA | Median %CV for the Processed Signal |
| gNegCtrlAveBGSubSig | 5.00 | -10.00 | Average of background subtracted signal of all inlier negative controls (BGSubSignal is calculated by substracting a value called BGUsed from the feature mean signal) |
| gNegCtrlAveNetSig | 40.00 | NA | Average of net signal of all inlier negative controls |
| gNegCtrlSDevBGSubSig | 10.00 | NA | Standard deviation of background subtracted signals of all inlier negative controls |
| gNonCntrlMedCVProcSignal | 8.00 | NA | Median %CV for the Processed Signal of the non-control probes |
| gSpatialDetrendRMSFilter | 15.00 | NA | Residual of background detrending fit |
Table 4: Principal parameters to be checked to verify the quality of microarray hybridization.
- Process the Microarray Data.
- Once all the microarray slides have been scanned and the quality control has been assessed positively, prepare a tab-delimited data-matrix by selecting the gProcessedSignal values from each single-subarray outcome file, representing the raw fluorescence intensities of each probe.
- In a spreadsheet, normalize the data on the 75th percentile within each array (P values) and calculate the average of all the P values among all different sub-arrays to calculate the R ratio.
- Then, on the same spreadsheet, normalize each gProcessedSignal value to the R ratio of its own sub-array.
4. Carry Out the Detailed Statistical Analysis of the Metabolomics and Transcriptomics Data
- Prepare the Statistical Analysis Software.
NOTE: In this case study, a software able to perform PCA, PLS-DA and O2PLS-DA was used.- Import the metabolomics and transcriptomics data. Go to File → New Regular Project → New Regular Project to import the data matrix obtained with MZmine software. Then click on Edit → Transpose the whole matrix → Home and assign the appropriate Primary and Secondary ID → Finish.
- Mean center the data and scale them using the pareto scale. In Home window, go to Edit → M1 and change the appropriate parameters depending the data. For the results described here, change the Scale from Unit Variance to Par.
- Scale the transcriptomics data using the Unit Variance setting.
- Carry Out the Multivariate Statistical Analysis.
- Implement the PCA as shown in Figure 2. In this case study, PCA reveals the main differences between samples, reflecting the different ripening stages and growing seasons.
- In the Workset window, select PCA-X as model type. Press Autofit. Pay attention to the R2X(cum) and Q2(cum) values as they give an idea about the quality of the model.
NOTE: Generally, the higher the values the better the model, but models with very high R2X(cum) might over-fit the data. As empirical rule, we stop adding Principal Components when the Q2(cum) value starts to decrease. - Then, select Scores → Scatter to see the plot that shows the possible grouping of the samples.
- Inspect the PCA Score Plot. If a good model is obtained (Q2cum >0.5), use the same classes of samples to build a O2PLS-DA model (step 4.2.2).
- In the Workset window, select PCA-X as model type. Press Autofit. Pay attention to the R2X(cum) and Q2(cum) values as they give an idea about the quality of the model.
- Build two O2PLS-DA models using the samples classified by macro-zone and validate the models using a permutation test with 200 permutations.
- Assign the classes by going to Home window → New As → M1 → Observations. Set the desired classes. Then, change the Model Type from PCA-X to O2PLS-DA. Press Autofit. Ensure that the number of components of the PLS-DA model is the same as that of the O2PLS-DA.
- To validate the O2PLS-DA model, go to Analyze CV-ANOVA and see the right p-value. Moreover, click on New As in the Home window → M2 and change the Model Type from O2PLS-DA to PLS-DA. Press Autofit.
- Go to Analyze → Permutations and perform 200 permutations (de-select Recalculate Permutations option).
Note: The final output shows a window in which the R2 value should generally hit the Y-axis in values under 0,4, while the Q2 should hit the negative part of the Y-axis. If R2 and/or Q2 values are incorrect, reduce the number of components both in PLS-DA and O2PLS-DA models. - Proceed to select M2 in Project Window and Click on Scores → Scatter to see the plot and the position of the sample classes. To observe what metabolites characterize one or more specific classes, go to Plot/List → Scatter.
- Change the Select Data Type to Observations and Loadings → Add Series and modify the Item in X-Axis and Series into pq(corr) and Pred Comp into 1 and either 2 or further components when present, respectively.
- With the mouse's right button pressed on the plot, go to Property → Color and select By Terms to distinguish the plot symbols among molecules and classes. Go to Layout → Format Plot → Axis and/or Styles to modify the plot with the desired characteristics.
- Based on the results of the metabolomics O2PLS-DA analysis, present differences in the relative levels of specific metabolites or classes of metabolites as histograms.
- Based on the results of the transcriptomics O2PLS-DA analysis, retrieve and assign differentially modulated genes to Gene Ontology (GO) classifications28.
- Identify relationships between metabolite levels and gene expression manually.
- Implement the PCA as shown in Figure 2. In this case study, PCA reveals the main differences between samples, reflecting the different ripening stages and growing seasons.
Wyniki
The case study described in this article yielded a final data matrix comprising 552 signals (m/z features) including molecular ions plus their isotopes, adducts and some fragments, relatively quantified among 189 samples (7 vineyards x 3 ripening stages x 3 growing seasons x 3 biological replicates). The total number for data points was therefore 104,328. Fragmentation tree analysis resulted in the annotation of 282 m/z features, corresponding to metabolites plus adducts...
Dyskusje
This article describes the metabolomics, transcriptomics and statistical analysis protocols used to interpret the grape berry terroir concept. Metabolomics analysis by HPLC-ESI-MS is sensitive enough to detect large numbers of metabolites simultaneously, but relative quantitation is affected by the matrix effect and ion suppression/enhancement. However, a similar approach has already been used to describe the ripening and post-harvest withering of Corvina berries, and the correction of matrix effects had a limited impact...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
This work benefited from the networking activities coordinated within the EU-funded COST ACTION FA1106 "An integrated systems approach to determine the developmental mechanisms controlling fleshy fruit quality in tomato and grapevine". This work was supported by the 'Completamento del Centro di Genomica Funzionale Vegetale' project funded by the CARIVERONA Bank Foundation and by the 'Valorizzazione dei Principali Vitigni Autoctoni Italiani e dei loro Terroir (Vigneto)' project funded by the Italian Ministry of Agricultural and Forestry Policies. SDS was financed by the Italian Ministry of University and Research FIRB RBFR13GHC5 project "The Epigenomic Plasticity of Grapevine in Genotype per Environment Interactions".
Materiały
| Name | Company | Catalog Number | Comments |
| Mill Grinder | IKA | IKA A11 basic | |
| HPLC Autosampler | Beckman Coulter | - | System Gold 508 Autosampler |
| HPLC System | Beckman Coulter | - | System Gold 127 Solvent Module HPLC |
| C18 Guard Column | Grace | - | Alltima HP C18 (7.5 mm x 2.1 mm; 5 μm) Guard Column |
| C18 Column | Grace | - | Alltima HP C18 (150 mm x 2.1 mm; 3 μm) Column |
| Mass Spectometer | Bruker Daltonics | - | Bruker Esquire 6000; The mass spectometer was equipped with an ESI source and the analyzer was an ion trap. |
| Extraction solvents and HPLC buffers | Sigma | 34966 | Methanol LC-MS grade |
| Sigma | 94318 | Formic acid LC-MS grade | |
| Sigma | 34967 | Acetonitrile LC-MS grade | |
| Sigma | 39253 | Water LC-MS grade | |
| Minisart RC 4 Syringe filters (0.2 μm) | Sartorius | 17764 | |
| Softwares for data collection (a) and processing (b) | Bruker Daltonics | - | Bruker Daltonics Esquire 5.2 Control (a); Esquire 3.2 Data Analysis and MzMine 2.2 softwares (b) |
| Spectrum Plant Total RNA kit | Sigma-Aldrich | STRN250-1KT | For total RNA extractino from grape pericarps |
| Nanodrop 1000 | Thermo Scientific | 1000 | |
| BioAnalyzer 2100 | Agilent Technologies | G2939A | |
| RNA 6000 Nano Reagents | Agilent Technologies | 5067-1511 | |
| RNA Chips | Agilent Technologies | 5067-1511 | |
| Agilent Gene Expression Wash Buffer 1 | Agilent Technologies | 5188-5325 | |
| Agilent Gene Expression Wash Buffer 2 | Agilent Technologies | 5188-5326 | |
| LowInput QuickAmp Labeling kit One-Color | Agilent Technologies | 5190-2305 | |
| Kit RNA Spike In - One-Color | Agilent Technologies | 5188-5282 | |
| Gene Expression Hybridization Kit | Agilent Technologies | 5188-5242 | |
| RNeasy Mini Kit (50) | Qiagen | 74104 | For cRNA Purification |
| Agilent SurePrint HD 4X44K 60-mer Microarray | Agilent Technologies | G2514F-048771 | |
| eArray | Agilent Technologies | - | https://earray.chem.agilent.com/earray/ |
| Gasket slides | Agilent Technologies | G2534-60012 | Enable Agilent SurePrint Microarray 4-array Hybridization |
| Thermostatic bath | Julabo | - | |
| Hybridization Chamber | Agilent Technologies | G2534-60001 | |
| Microarray Hybridization Oven | Agilent Technologies | G2545A | |
| Hybridization Oven Rotator Rack | Agilent Technologies | G2530-60029 | |
| Rotator Rack Conversion Rod | Agilent Technologies | G2530-60030 | |
| Staining kit | Bio-Optica | 10-2000 | Slide-staining dish and Slide rack |
| Magnetic stirrer device | AREX Heating Magnetic Stirrer | F20540163 | |
| Thermostatic Oven | Thermo Scientific | Heraeus - 6030 | |
| Agilent Microarray Scanner | Agilent Technologies | G2565CA | |
| Scanner Carousel, 48-position | Agilent Technologies | G2505-60502 | |
| Slide Holders | Agilent Technologies | G2505-60525 | |
| Feature extraction software v11.5 | Agilent Technologies | - | inside the Agilent Microarray Scanner G2565CA |
| SIMCA + V13 Software | Umetrics |
Odniesienia
- Jessome, L. L., Volmer, D. A. Ion suppression: A major concern in mass spectrometry. Lc Gc N Am. 24 (5), 498-510 (2006).
- Kim, H. K., Choi, Y. H., Verpoorte, R. NMR-based plant metabolomics: where do we stand, where do we go?. Trends Biotech. 29 (6), 267-275 (2011).
- Sumner, L. W., Mendes, P., Dixon, R. A. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochem. 62 (6), 817-836 (2003).
- Bottcher, C., von Roepenack-Lahaye, E., Willscher, E., Scheel, D., Clemens, S. Evaluation of matrix effects in metabolite profiling based on capillary liquid chromatography electrospray ionization quadrupole time-of-flight mass spectrometry. Anal Chem. 79 (4), 1507-1513 (2007).
- Toffali, K., et al. Novel aspects of grape berry ripening and post-harvest withering revealed by untargeted LC-ESI-MS metabolomics analysis. Metabolomics. 7 (3), 424-436 (2011).
- Martin, J. C., et al. Can we trust untargeted metabolomics? Results of the metabo-ring initiative, a large-scale, multi-instrument inter-laboratory study. Metabolomics. 11 (4), 807-821 (2015).
- Jaillon, O., et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 449 (7161), 463-467 (2007).
- Velasco, R., et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. Plos One. 2 (12), (2007).
- Tornielli, G. B., Zamboni, A., Zenoni, S., Delledonne, M., Pezzotti, M., Gerós, H., Chaves, M., Delrot, S. Ch. 11. The Biochemestry of the Grape Berry. 11, (2012).
- Anesi, A., et al. Towards a scientific interpretation of the terroir concept: plasticity of the grape berry metabolome. BMC Plant Biol. 15, 1-17 (2015).
- Berdeja, M., et al. Water limitation and rootstock genotype interact to alter grape berry metabolism through transcriptome reprogramming. Hort Res. 2, 1-13 (2015).
- Carbonell-Bejerano, P., et al. Solar ultraviolet radiation is necessary to enhance grapevine fruit ripening transcriptional and phenolic responses. BMC Plant Biol. 14, 1-16 (2014).
- Carbonell-Bejerano, P., et al. Reducing sampling bias in molecular studies of grapevine fruit ripening: transcriptomic assessment of the density sorting method. Theor Exp Plant Phys. 28 (1), 109-129 (2016).
- Carbonell-Bejerano, P., et al. Circadian oscillatory transcriptional programs in grapevine ripening fruits. BMC Plant Biol. 14, 1-15 (2014).
- Cavallini, E., et al. Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant & Cell Physiol. 55 (3), 517-534 (2014).
- Cramer, G. R., et al. Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol. 14, 1-21 (2014).
- Dal Santo, S., et al. The plasticity of the grapevine berry transcriptome. Genome Biol. 14 (6), 1-17 (2013).
- Fasoli, M., et al. The Grapevine Expression Atlas Reveals a Deep Transcriptome Shift Driving the Entire Plant into a Maturation Program. Plant Cell. 24 (9), 3489-3505 (2012).
- Gambino, G., et al. Co-evolution between Grapevine rupestris stem pitting-associated virus and Vitis vinifera L. leads to decreased defence responses and increased transcription of genes related to photosynthesis. J Exp Bot. 63 (16), 5919-5933 (2012).
- Ghan, R., et al. Five omic technologies are concordant in differentiating the biochemical characteristics of the berries of five grapevine (Vitis vinifera L.) cultivars. BMC Genomics. 16 (1), 1-26 (2015).
- Pastore, C., et al. Selective defoliation affects plant growth, fruit transcriptional ripening program and flavonoid metabolism in grapevine. BMC Plant Biol. 13, 1-13 (2013).
- Pastore, C., et al. Increasing the source/sink ratio in Vitis vinifera (cv Sangiovese) induces extensive transcriptome reprogramming and modifies berry ripening. BMC Genomics. 12, 1-23 (2011).
- Rinaldo, A. R., et al. A Grapevine Anthocyanin Acyltransferase, Transcriptionally Regulated by VvMYBA, Can Produce Most Acylated Anthocyanins Present in Grape Skins. Plant Physiol. 169 (3), 1897-1916 (2015).
- Royo, C., et al. Developmental, transcriptome, and genetic alterations associated with parthenocarpy in the grapevine seedless somatic variant Corinto bianco. J Exp Bot. , 259-273 (2015).
- Venturini, L., et al. De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics. 14, 1-13 (2013).
- Commisso, M., Strazzer, P., Toffali, K., Stocchero, M., Guzzo, F. Untargeted metabolomics: an emerging approach to determine the composition of herbal products. Comput Struct Biotechnol J. 4, 1-7 (2013).
- Pluskal, T., Castillo, S., Villar-Briones, A., Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 11, 1-11 (2010).
- Ashburner, M., et al. Gene Ontology: tool for the unification of biology. Nat Genet. 25 (1), 25-29 (2000).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone