Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Simultaneous Study of the Recruitment of Monocyte Subpopulations Under Flow In Vitro
W tym Artykule
Podsumowanie
Here, we present an integrated protocol that measures monocyte subpopulation trafficking under flow in vitro by use of specific surface markers and confocal fluorescence microscopy. This protocol can be used to explore sequential recruitment steps as well as to profile other leukocyte subtypes using other specific surface markers.
Streszczenie
The recruitment of monocytes from the blood to targeted peripheral tissues is critical to the inflammatory process during tissue injury, tumor development and autoimmune diseases. This is facilitated through a process of capture from free flow onto the luminal surface of activated endothelial cells, followed by their adhesion and transendothelial migration (transmigration) into the underlying affected tissue. However, the mechanisms that support the preferential and context-dependent recruitment of monocyte subpopulations are still not fully understood. Therefore, we have developed a method that allows the recruitment of different monocyte subpopulations to be simultaneously visualized and measured under flow. This method, based on time-lapse confocal imaging, allows for the unambiguous distinction between adherent and transmigrated monocytes. Here, we describe how this method can be used to simultaneously study the recruitment cascade of pro-angiogenic and non-angiogenic monocytes in vitro. Furthermore, this method can be extended to study the different steps of recruitment of up to three monocyte populations.
Wprowadzenie
Monocytes constitute a phagocytic component of innate immunity that is essential for fighting pathogens, cleaning up damaged tissues, angiogenesis, and the pathophysiology of many diseases including cancers1,2,3. Monocytes are bone marrow-derived cells composed of heterogeneous subpopulations that circulate in the blood but can be recruited to the site of inflammation in peripheral tissue through specific molecular mechanisms. The recruitment cascades of monocytes, as for leukocytes in general, implicates different steps including capture, rolling, crawling, arrest, transendothelial migration (transmigration) and migration through the vessel wall (basement membrane and mural cells)4. These steps mainly involve inflammation-induced molecules on the endothelial luminal surface such as selectins, glycoprotein ligands, chemokines, intercellular and junctional adhesion molecules, and their receptors on leukocytes such as selectin ligands and integrins. Trafficking pathways through either the endothelial cell junctions (paracellular) or through the endothelial cell body (transcellular) can be used by leukocytes to cross the endothelial barrier5. Whilst monocytes have historically been documented to transmigrate through the transcellular route, potential divergences in their migratory pathway have been proposed as monocytes are no longer considered a homogeneous cell population. It is now becoming clear that monocyte diversity can be defined by each of their differences and commonalities, with respect to their distinctive extravasation cascades3,6. Therefore, in order to unambiguously discriminate between monocyte subpopulations, it is crucial to visualize and phenotype the behavior of each of these different subpopulations during the recruitment process.
Monocytes from human, pig, rat and mouse were subdivided into phenotypic subpopulations with certain ascribed functions and specific migratory behaviors7,8,9. For example, in humans, monocytes can be divided into three subsets based on their surface expression of CD14, a coreceptor for bacterial lipopolysaccharide, and CD16, the Fc-gamma receptor III. Human monocyte subpopulations include classical CD14+CD16-, intermediate CD14+CD16+ and non-classical CD14dimCD16+ cells6,9. The classical CD14+CD16- monocytes were shown to be mainly inflammatory whereas the pool of CD16+ monocytes were collectively found to present TIE2 expression and proangiogenic function10. Consistently, endothelial cell stimulation with inflammatory cytokines such as human tumor necrosis factor (TNF)α or interleukin (IL-1)beta (conventional inflammation) is sufficient to trigger the complete recruitment of classical CD14+CD16- monocytes. However, simultaneous actions of vascular endothelial growth factor (VEGF)A and TNFα (angiogenic factors-driven inflammation) are required to provoke the transmigration of the CD16+ proangiogenic pool of monocytes3. Historically, the traditional Transwell system under static conditions, the parallel plate flow chamber, and the µ-slide flow chambers have been used to quantitatively analyze the recruitment of one leukocyte population at a time in vitro11,12,13. Whilst these protocols have been validated, a more robust method that allowed the simultaneous analysis of multiple monocyte subpopulations would be considered more insightful. Such methodologies must account for multiple interactions and the differing frequencies of each respective population and also provide a mechanistic understanding of the similarities and specificities for the recruitment cascades that define each monocyte subset.
Here, we present a method based on the time-lapse imaging of monocyte recruitment under flow which allows the migratory cascades of different monocyte subpopulations to be studied simultaneously by using confocal microscopy. This method integrates certain critical features that mimic endothelial cell inflammation, as well as the hemodynamics of circulating monocytes in post-capillary venules, the main location of leukocyte recruitment in vivo. The proposed method uses human umbilical vein endothelial cells (HUVEC), which are generated through a well-established protocol of isolation from human umbilical cords. This clinical resource has the advantage of being easily available as a biological by-product, whilst also providing a reasonable yield of endothelial cells that can be isolated from the umbilical vein. We also used fluorescent dyes and immunofluorescence to distinguish between the different cellular components, and confocal microscopy to unambiguously define monocyte positioning (luminal vs. abluminal) over time. The protocol presented here has been developed to simultaneously measure the transmigration levels of monocyte subpopulations. Moreover, it should be noted that this methodology can be extended to study other leukocytes subpopulations and recruitment processes by use of different biomarkers and labelling.
Protokół
Human materials were used with the informed consent of volunteer donors and in accordance with the Swiss Ethics Committees on clinical research.
1. Isolation and Freezing of Human Umbilical Vein Endothelial Cells (HUVEC)
- Add 5 mL of coating solution to a T75 flask (0.1 mg/mL collagen G and 0.2% gelatin in phosphate buffered saline PBS at pH 7.4) for 30 min at 37 °C before initiating HUVEC isolation.
- Clean the cord with PBS, wipe it with sterile compresses, and place it in a sterile 20 cm Petri dish. Cut the ends of the cord with sterile scissors.
- Identify the single large vein and the two small arteries. Gently insert a cannula with a three-way stopcock attached to it into the vein extremities at the cord ends.
- Tighten the cord and the cannula connection firmly with a length of wire.
- Perfuse the cord twice with 20 mL of RPMI medium containing 100 U/mL penicillin, 100 U/mL streptomycin and 250 ng/mL amphotericin B to wash the cord’s veins. This process makes the appearance of the cord whiter and clearer. Empty the vein before collagenase addition by collecting the RPMI with a syringe at one end.
- Perfuse the vein with 12 mL of 1 mg/mL collagenase type I (0.22 µm-filtered).
- Close the stopcock at the cord ends and incubate the cord at 37 °C for 12 min.
- Gently massage the cord to detach endothelial cells from the vein lumen.
- Take 30 mL of RPMI containing 10% fetal calf serum with a 50 mL syringe and connect it to one end of the umbilical cord.
- Connect an empty 50 mL syringe to the other end of the umbilical cord
- Open the stopcock and perfuse the vein from one end whilst reciprocally collecting from the other end.
NOTE: The collected suspension contains endothelial cells. - Centrifuge this cell suspension at 200 x g for 5 min.
- Discard the supernatant and resuspend the cell pellet with 10 mL of complete M199 medium (M199 containing 20% FCS, 15 µg/mL endothelial cell growth supplements, 100 µg/mL heparin sodium, 0.5 µM hydrocortisone, 10 µg/mL L-Ascorbic Acid, 100 U/mL penicillin, 100 U/mL streptomycin and 250 ng/mL amphotericin B).
- Remove the coating solution from the T75 flask and rinse once with PBS.
- Seed the cells collected from step 1.13 into the T75 flask and place it in the incubator at 37 °C with 5% CO2.
- The next day, rinse the flask 3 times with the complete M199 medium to remove residual red blood cells and then change the medium every 2 days until confluence.
- At 80–90% confluence, rinse the HUVEC monolayer once with 5 mL of PBS and detach the cells with 5 mL of 0.05% trypsin in 1 mM EDTA at 37 °C for 5 min. Add 4 mL of M199 and 1 mL of FCS to stop the trypsin action. Flush the flask to detach all HUVEC.
- Collect an aliquot of 50 µL to be used for staining of VE-cadherin, PECAM-1 and gp38, and analyze by flow cytometry to check HUVEC purity.
- Collect the remainder of HUVEC from step 1.18 in a 15 mL tube and centrifuge at 200 x g for 5 min at room temperature.
- Discard the supernatant from step 1.19, resuspend the cell pellet in freezing solution (FCS containing 10% DMSO) at a density of 5 x 105 cells/mL in cryotubes, and freeze at -80 °C or in liquid nitrogen until use.
- To check HUVEC purity:
- Add 1 µL of anti-human VE-cadherin-FITC antibody, 1 µL of anti-human PECAM1-PE antibody, and 1 µL of anti-human Podoplanin-APC antibody to the aliquot of 50 µL of HUVEC collected at step 1.18.
- Incubate at room temperature for 10 min.
- Add 100 µL of PBS and centrifuge at 400 x g for 30 s.
- Discard the supernatant and resuspend in 100 µL of PBS. Data can now be acquired by flow cytometry techniques.
NOTE: HUVEC are positive for VE-cadherin and PECAM-1, and negative for Podoplanin.
2. HUVEC Defrosting
NOTE: Use HUVEC at low passage for experiments (maximum 5 passages).
- Coat a T75 flask with 1 mL of the coating solution at 37 °C for 30 min.
- Rapidly defreeze HUVEC at 37 °C for 2 min and resuspend the cells in 10 mL of complete M199.
- Centrifuge the cells at 200 x g at room temperature for 5 min and discard the supernatant.
- Resuspend the cell pellet in 10 mL of complete M199.
- Transfer the cell suspension in the pre-coated flask. Place the flask in the incubator at 37 °C with 5% CO2. Change the cell culture medium every 2 days.
3. HUVEC Culture in 0.4 µ-Slide Chamber
- Five days before starting the flow experiment, pre-coat the chambers of a 0.4 µ-slide with 30 µL of PBS containing 0.1 mg/mL collagen G, 0.2% gelatin at 37 °C for 30 min.
- Wash the chambers with 100 µL of PBS.
- Detach the cells from an 80–90% confluent HUVEC of a T75 flask.
- Rinse HUVEC with 5 mL of PBS and detach them with 5 mL of 0.05% trypsin at 37 °C for 5 min.
- Flush and collect the cell suspension in complete M199 and count the cells by the most convenient method. Centrifuge at 200 x g for 5 min at room temperature.
- Resuspend the cell pellet at 106 cells/mL and distribute 30 µL (30,000 cells) per chamber.
- Incubate the cells in an incubator at 37 °C with 5% CO2 for 1 h.
- Add 150 µL of complete M199 to each chamber and culture the cells for 5 days in the incubator at 37 °C and 5% CO2. Change the medium every 2 days.
4. HUVEC Staining for Monocyte Recruitment Assay Under Flow
- Prepare the labeling medium made of M199 and 1 µM of CMFDA (5-chloromethylfluorescein diacetate) and warm it at 37 °C for 5 min before cell labeling.
- Wash HUVEC twice with M199 medium warmed at 37 °C.
- Replace the medium with 30 µL of warmed labeling medium containing 1 µM of CMFDA and place into the incubator at 37 °C and 5% CO2 for 10 min.
- Wash once with complete M199 and incubate the cells with complete M199 in the incubator at 37 °C and 5% CO2 for 30 min.
NOTE: It is important to remove all traces of serum before addition of the labeling solution, otherwise it may alter HUVEC staining. - Replace the medium with complete M199 containing either human TNFα (500 U/mL) or a mix of human TNFα (500 U/mL) with human VEGFA (1 µg/mL) for 6 h in an incubator at 37 °C and 5% CO2.
5. Isolation of Human Pan Monocytes and Staining of Subpopulations
- Use either a buffy coat of concentrated human blood, or 20 mL of freshly isolated human blood, collected on the day of the experiment in EDTA vacutainer tubes.
- Dilute the blood in PBS-1 mM EDTA (1:1) and pipette gently 20 mL of the diluted blood on top of the 20 mL of density gradient media. Centrifuge at 400 x g for 30 min at room temperature with slow acceleration and without brake.
- Collect the peripheral blood mononuclear cell (PBMC)-platelet layer (between density gradient media and plasma layers) into a new 50 mL tube containing 40 mL of PBS- 1 mM EDTA. Top up to 50 mL with PBS- 1 mM EDTA.
- Centrifuge at 200 x g at room temperature for 5 min. Discard the supernatant.
- Resuspend the cell pellet with 10 mL of staining buffer (PBS- 1 mM EDTA containing 0.5% bovine serum albumin BSA).
- Centrifuge at 200 x g at room temperature for 5 min. Discard the supernatant.
- Repeat steps 5.5 and 5.6.
- Resuspend the cell pellet with 10 mL of staining buffer. Take an aliquot of 10 µL for a cell count.
- Check PBMC populations and count cells rapidly with a flow cytometer.
NOTE: The characteristic lymphocyte and monocyte populations can be observed (Figure 1A). From 50 mL of fresh human blood expect about 50-100 x 106 PBMC. - For the recruitment of CD14+ versus CD14- PBMC under flow:
- Wash the pellet three times with flow buffer (M199 containing 0.5% BSA) and resuspend the mononuclear cells in flow buffer at 6 x 106 cells per mL.
- Make aliquots of 200 µL for each assay. Incubate at 37 °C until 20 min before the assay.
- Add 5 µL of anti-CD14-PE and Hoechst 33342 at a final concentration of 2 µM to each aliquot. Mix and incubate at 37 °C for 10 min.
- Centrifuge the aliquot at 400 x g for 30 s.
- Discard the supernatant and resuspend the pellet with 200 µL of flow buffer.
- For the recruitment of monocyte subpopulations under flow:
- Isolate monocytes with a pan monocyte isolation kit according to manufacturer instructions.
NOTE: The following isolation protocol is for 50 x 106 cells. It can be scaled up or down as long as it is within the manufacturer’s recommendations. - Centrifuge the PBMC suspension at 200 x g at room temperature for 5 min.
- Discard the supernatant and resuspend the pellet with 400 µL of staining buffer.
- Add 50 µL of Fc-receptor blocking reagent and 50 µL of Pan Monocyte antibody cocktail.
- Incubate at room temperature for 10 min.
- Add 400 µL of staining buffer and 100 µL of magnetic beads conjugated anti-biotin antibody. Incubate at room temperature for 15 min.
- Add 2 mL of staining buffer and use a MACS LS column coupled with a magnet.
- Place the LS column on the magnet and add 1 mL of staining buffer. Discard the flow-through.
- Pass the PBMC suspension in the column and collect the clear flow though containing pan monocytes in a new 15 mL tube.
- Add the staining buffer to top up to 5 mL.
- Take an aliquot and check the quality of the monocyte isolation with a flow cytometer.
- Determine the pan monocyte count.
NOTE: Only monocyte population can be observed (Figure 1B). - Centrifuge the remainder of monocytes from step 5.11.11 at 200 x g for 5 min.
- Discard the supernatant.
- Resuspend the cell pellet in 5 mL of flow buffer (M199 containing 0.5% BSA).
- Repeat 5.11.13 to 5.11.14 twice to eliminate any trace of EDTA.
- Isolate monocytes with a pan monocyte isolation kit according to manufacturer instructions.
- Make monocyte suspension in flow buffer (M199 with 0.5% BSA) at 6 x 106 cells/mL.
- Make aliquots of 200 µL of monocytes for each recruitment assay.
- Keep the aliquot at 37 °C in the incubator until 20 min before injection.
- Add 5 µL of anti-CD16-PE antibody and Hoechst 33342 (2 µM final) to each aliquot.
- Mix and incubate at 37 °C for 10 min.
- Centrifuge the aliquot at 400 x g for 30 s.
- Discard the supernatant and resuspend the pellet with 250 µL of flow buffer.
- Add 30 µL of the monocyte suspension in one chamber of the slide to serve for setting the acquisition parameters on the confocal microscope.
- Keep the aliquots of monocyte suspension from step 5.18 at 37 °C.
NOTE: This suspension is ready to be injected in the flow system.
6. Preparation of the Fluidic System
- Ensure that the cell incubator for the imaging set at 37 °C.
NOTE: A diagram of the flow system is shown in Figure 2. - Assemble the tubing part I: Insert a Luer connector male to one end of a piece of silicone tubing (8 cm long and 3 mm thick) and connect the other end to an in-line Luer injection set. Connect the latter Luer connector to a piece of silicone tubing (40 cm and 3 mm thick) at one end.
NOTE: Optionally, a 3-way tap connected to a 5 mL syringe can be inserted between the in-line Luer injection set and the silicone tubing for eventual air bubble removal. - Assemble the tubing part II: Connect a 20 mL syringe to one end of a length of silicone tubing (1 m long and 3 mm thick). Insert a Luer connector male to the other end of the tubing.
- Connect part I and part II tubing by inserting the Luer connector males to a female Luer lock coupler (Figure 2A).
- Put the free end of the silicone tubing in the reservoir containing the flow buffer (M199 + 0.5% BSA) warmed at 37 °C.
- Pull on the plunger of the 20 mL syringe to fill the tubing with flow buffer.
- Place the syringe on the pump and secure it.
- Set the pump in withdraw mode (as opposed to infuse) and specify the flow rate.
- Determine the flow rate according to the IBIDI slide used by using the following formula:
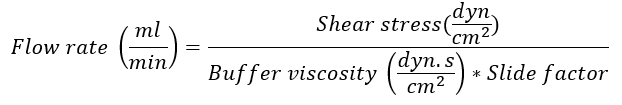
Note: The slide factor is dependent on the IBIDI slide used for the experiment. For the µ-slide I0.4 Luer lock used in this example, the slide factor is 131.6. For specific slide factors, see the company website14. The flow buffer viscosity is 0.0072 dyn.s/cm2. Shear stress at the post-capillary venules is about 0.5 dyn/cm2. - Connect the slide (Figure 2B):
- Clamp the silicone tubing around the female Luer Lock Coupler and disconnect the two Luer connector males from the coupler.
- Connect them to the reservoirs of the slide containing stimulated HUVEC and fill with medium. Avoid air bubbles during this step.
- Take off the clamps and ensure that the connection is not leaking.
- Place the slide under the microscope for time-lapse imaging and start the pump.
7. Time-lapse Imaging of Monocyte Recruitment Under Flow by Confocal Microscopy
- Use a 40x objective (see Table of Materials) for imaging.
- Activate the 405 nm (blue monocyte nuclei), 488 nm (green endothelial cells) and 561 nm (red CD16+ subset) lasers.
- Use the chamber that contains the monocytes to set the acquisition parameters.
NOTE: To detect both non-transmigrated and transmigrated monocytes, the pinhole and intensity of the laser 405 nm are set high. Thus, non-transmigrated monocytes are slightly visible in the basal plan. However only transmigrated monocytes present an unstained area around the nucleus corresponding to the new space occupied underneath endothelial cells. - Place the chamber to be acquired under the microscope.
- Choose 3 fields of views within 1 cm radius for multi-position confocal imaging.
- Define the basal and the apical sides of endothelial cells
- Set a z-stack to the 10–12 µm range (0.5 µm step). Run a time-lapse acquisition every 1 min.
- After 3 min of imaging, inject 200 µL of monocyte suspension (6x106 cells/mL) through the in-line Luer injection port.
NOTE: Rapidly monocytes appear in the apical focal plane, adhere and start transmigration (transit from the apical to the basal plan). - Image for at least 30 min. Once finished, stop imaging and stop the flow. Clamp the tubing to disconnect them from the slide.
- Fix the slide with 4% paraformaldehyde at 4 °C for 10 min.
- Wash the slide with PBS and store the slide at 4 °C for further analysis if needed.
8. Analyze the Data with ImageJ
- Count the number of total adherent monocytes in each field. Determine the cell count per mm2.
- Count transmigrated monocytes that are present in the basal plan underneath endothelial cells and identified by the presence of a black hole (in the green channel) around the nucleus.
- Divide the count of transmigrated leukocytes by the total number of adherent leukocytes. The transmigration rate is presented as a percentage of adherent monocytes.
- For illustration, the apical and the basal sides can be shown simultaneously to illustrate the events occurring in each of these endothelial compartments.
Wyniki
Determining the state of HUVEC activation induced by TNFα
The bio-activity of the inflammatory cytokine TNFα can be vary according to the batch and the repletion of freezing-thawing cycle. It is important to check the activation status of HUVEC with TNFα treatment. This could be performed by staining in parallel some samples of confluent HUVEC for the inflammatory induction of selectins, ICAM-1 and VCAM-1
Dyskusje
Here, we report a method detailing a study of how monocyte subpopulations transmigrate through the inflamed endothelial monolayer. The discussed method used confocal microscopy instead of phase-contrast microscopy, which is also used to study monocyte recruitment under flow3,11,19. One major advantage of using confocal microscopy for time-lapse imaging is the ability to unambiguously discriminate between transmigration and stron...
Ujawnienia
The authors have no competing financial interests.
Podziękowania
We thank Dr. Paul Bradfield for manuscript reading and feedbacks. A. S. received financial support from the Sir Jules Thorn Charitable Overseas Trust Reg.,
Materiały
| Name | Company | Catalog Number | Comments |
| Tissue Culture Flasks 75 cm2 | TPP | 90076 | Routine culture of isolated HUVEC |
| µ-Slide VI 0.4 | IBIDI | 80606 | |
| Centrifuge Tubes 15 mL | TPP | 191015 | |
| Centrifuge Tubes 50 mL | TPP | 191050 | |
| Collagen G | Biochrom | L 7213 | For coating of cell culture flasks |
| Gelatin | Sigma-Aldrich | 1393 | For coating of cell culture flasks |
| Dulbecco’s Phosphate Buffered Saline (without MgCl2 and CaCl2) | Sigma-Aldrich | D8537 | |
| Dulbecco’s Phosphate Buffered Saline (with MgCl2 and CaCl2) | Sigma-Aldrich | D8662 | |
| RPMI-1640 Medium | Sigma-Aldrich | R8758 | |
| 3-Way Stopcocks | BIO-RAD | 7328103 | |
| penicillin 10,000 μ/mL streptomycine 10,000 μg/mL fungizone 25 μ/mL | AMIMED | 4-02F00-H | |
| Collagenase type 1 | Worthington | LS004216 | |
| Medium 199 1x avec Earle's salts, L-Glutamine, 25 mM Hepes | GIBCO | 22340020 | |
| Bovine Albumin Fraction V | ThermoFisher | 15260037 | |
| Endothelial Cell Growth Supplement, 150 mg | Millipore | 02-102 | |
| Heparin Sodium | Sigma-Aldrich | H3149RT | |
| Hydrocortisone | Sigma-Aldrich | H6909 | |
| L-Ascorbic acid | Sigma-Aldrich | A 4544 | |
| EDTA disodium salt dihydrate C10H14N2Na2O8 · 2H2O | APPLICHEM | A2937.0500 | |
| CD144 (VE-Cadherin), human recombinant clone: REA199, FITC | Miltenyi Biotech | 130-100-713 | AB_2655150 |
| CD31-PE antibody, human recombinant clone: REA730, PE | Miltenyi Biotech | 130-110-807 | AB_2657280 |
| Anti-Podoplanin-APC, human recombinantclone: REA446, APC | Miltenyi Biotech | 130-107-016 | AB_2653263 |
| BD Accuri C6 Plus | BD Bioscience | ||
| µ-Slide I Luer | IBIDI | 80176 | |
| CMFDA (5-chloromethylfluorescein diacetate) | ThermoFisher | C2925 | |
| Recombinant human TNFα | Peprotech | 300-01A | |
| Recombinant human VEGFA | Peprotech | 100-20 | |
| NE-1000 Programmable Syringe Pump | KF Technology | NE-1000 | |
| Ficoll Paque Plus | GE Healthcare | 17-1440-02 | |
| Anti-human CD14-PE, human recombinant clone: REA599, PE | Miltenyi Biotech | 130-110-519 | AB_2655051 |
| Pan Monocyte Isolation Kit, human | Miltenyi Biotech | 130-096-537 | |
| Anti-human CD16-PE, human recombinant clone: REA423, PE | Miltenyi Biotech | 130-106-762 | AB_2655403 |
| LS columns | Miltenyi Biotech | 130-042-401 | |
| QuadroMACS Separator | Miltenyi Biotech | 130-090-976 | |
| Hoechst 33342, Trihydrochloride, Trihydrate | ThermoFisher | H1399 | |
| Silicone tubing | IBIDI | 10841 | |
| Elbow Luer Connector | IBIDI | 10802 | |
| Female Luer Lock Coupler | IBIDI | 10823 | |
| Luer Lock Connector Female | IBIDI | 10825 | |
| In-line Luer Injection Port | IBIDI | 10820 | |
| Ar1 confocal microscope | Nikon | ||
| 40x objective | Nikon | 40x 0.6 CFI ELWD S Plane Fluor WD:3.6-2.8mm correction 0-2mm | |
| ImageJ Software | NIH |
Odniesienia
- Auffray, C., Sieweke, M. H., Geissmann, F. Blood Monocytes: Development, Heterogeneity, and Relationship with Dendritic Cells. Annual Review of Immunology. 27 (1), 669-692 (2009).
- De Palma, M., Venneri, M. A., Roca, C., Naldini, L. Targeting exogenous genes to tumor angiogenesis by transplantation of genetically modified hematopoietic stem cells. Nature Medicine. 9 (6), 789-795 (2003).
- Sidibe, A., et al. Angiogenic factor-driven inflammation promotes extravasation of human proangiogenic monocytes to tumours. Nature Communications. 9 (1), 355 (2018).
- Ley, K., Laudanna, C., Cybulsky, M. I., Nourshargh, S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nature Review Immunology. 7 (9), 678-689 (2007).
- Nourshargh, S., Alon, R. Leukocyte Migration into Inflamed Tissues. Immunity. 41 (5), 694-707 (2014).
- Cros, J., et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 33 (3), 375-386 (2010).
- Geissmann, F., Jung, S., Littman, D. R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 19 (1), 71-82 (2003).
- Chamorro, S., et al. In vitro differentiation of porcine blood CD163− and CD163+ monocytes into functional dendritic cells. Immunobiology. 209 (1-2), 57-65 (2004).
- Passlick, B., Flieger, D., Ziegler-Heitbrock, H. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood. 74 (7), (1989).
- Venneri, M. A., et al. Identification of proangiogenic TIE2-expressing monocytes (TEMs) in human peripheral blood and cancer. Blood. 109 (12), 5276-5285 (2007).
- Bradfield, P. F., et al. JAM-C regulates unidirectional monocyte transendothelial migration in inflammation. Blood. 110 (7), 2545-2555 (2007).
- Schenkel, A. R., Mamdouh, Z., Muller, W. A. Locomotion of monocytes on endothelium is a critical step during extravasation. Nature Immunology. 5 (4), 393-400 (2004).
- Luu, N. T., Rainger, G. E., Nash, G. B. Kinetics of the different steps during neutrophil migration through cultured endothelial monolayers treated with tumour necrosis factor-alpha. Journal Vascular Research. 36 (6), 477-485 (1999).
- ibidi GmbH. . Shear Stress and Shear Rates for ibidi µ-Slides - Based on Numerical Calculations. , (2014).
- Yang, L., Froio, R. M., Sciuto, T. E., Dvorak, A. M., Alon, R., Luscinskas, F. W. ICAM-1 regulates neutrophil adhesion and transcellular migration of TNF-alpha-activated vascular endothelium under flow. Blood. 106 (2), 584-592 (2005).
- Yang, C. -. R., Hsieh, S. -. L., Ho, F. -. M., Lin, W. -. W. Decoy receptor 3 increases monocyte adhesion to endothelial cells via NF-kappa B-dependent up-regulation of intercellular adhesion molecule-1, VCAM-1, and IL-8 expression. Journal of Immunology. 174 (3), 1647-1656 (2005).
- Wong, D., Dorovini-Zis, K. Expression of vascular cell adhesion molecule-1 (VCAM-1) by human brain microvessel endothelial cells in primary culture. Microvascular Research. 49 (3), 325-339 (1995).
- Bradfield, P. F., Nourshargh, S., Aurrand-Lions, M., Imhof, B. A. JAM family and related proteins in leukocyte migration (Vestweber series). Arteriosclerosis Thrombosis and Vascular Biology. 27 (10), 2104-2112 (2007).
- Bradfield, P. F., et al. Divergent JAM-C Expression Accelerates Monocyte-Derived Cell Exit from Atherosclerotic Plaques. PLoS One. 11 (7), e0159679 (2016).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone