A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Site-specific Bacterial Chromosome Engineering: ΦC31 Integrase Mediated Cassette Exchange (IMCE)
In This Article
Summary
A quick and efficient method to integrate foreign DNA of interest into pre-made acceptor strains, termed landing pad strains, is described. The method allows site-specific integration of a DNA cassette into the engineered landing pad locus of a given strain, through conjugation and expression of the ΦC31 integrase.
Abstract
The bacterial chromosome may be used to stably maintain foreign DNA in the mega-base range1. Integration into the chromosome circumvents issues such as plasmid replication, plasmid stability, plasmid incompatibility, and plasmid copy number variance. This method uses the site-specific integrase from the Streptomyces phage (Φ) C312,3. The ΦC31 integrase catalyzes a direct recombination between two specific DNA sites: attB and attP (34 and 39 bp, respectively)4. This recombination is stable and does not revert5. A "landing pad" (LP) sequence consisting of a spectinomycin- resistance gene, aadA (SpR), and the E. coli ß-glucuronidase gene (uidA) flanked by attP sites has been integrated into the chromosomes of Sinorhizobium meliloti, Ochrobactrum anthropi, and Agrobacterium tumefaciens in an intergenic region, the ampC locus, and the tetA locus, respectively. S. meliloti is used in this protocol. Mobilizable donor vectors containing attB sites flanking a stuffer red fluorescent protein (rfp) gene and an antibiotic resistance gene have also been constructed. In this example the gentamicin resistant plasmid pJH110 is used. The rfp gene6 may be replaced with a desired construct using SphI and PstI. Alternatively a synthetic construct flanked by attB sites may be sub-cloned into a mobilizable vector such as pK19mob7. The expression of the ΦC31 integrase gene (cloned from pHS628) is driven by the lac promoter, on a mobilizable broad host range plasmid pRK78139.
A tetraparental mating protocol is used to transfer the donor cassette into the LP strain thereby replacing the markers in the LP sequence with the donor cassette. These cells are trans-integrants. Trans-integrants are formed with a typical efficiency of 0.5%. Trans-integrants are typically found within the first 500-1,000 colonies screened by antibiotic sensitivity or blue-white screening using 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid (X-gluc). This protocol contains the mating and selection procedures for creating and isolating trans-integrants.
Protocol
1. Production of Culture
- Prepare sterile liquid media: TY10 (5 g/l tryptone, 3 g/l yeast extract, 0.44 g/l calcium chloride dehydrate), and LB11 (10 g/l tryptone, 5 g/l yeast extract, 5 g/ sodium chloride, pH 7).
- Inoculate from a single colony: SmUW227 (S. meliloti LP-strain: construction will be described elsewhere, strain construction details available upon request) () into 5 ml of TY media with 50 μg/ml spectinomycin. Inoculate the following strains into 5 ml of LB liquid media, supplementing with the given antibiotic: E. coli DH5α12 containing pJC2, the integrase expression plasmid, with 10 μg/ml tetracycline; E. coli MT61613, the mobilizer, with 10 μg/ml chloramphenicol; and E. coli DH5α containing pJH110, the donor cassette plasmid, with 5 μg/ml gentamicin.
- Incubate E. coli strains overnight at 37 °C with constant shaking. Incubate S. meliloti strain at 30 °C for two days with shaking. It is possible to obtain a S. meliloti culture overnight with a larger inoculum (1:500 subculture of a saturated culture).
2. Culture Preparation and Mixing
- Wash 1.5 ml of culture for each strain by collecting the cell pellet by centrifugation at 17,000 x g for 30 seconds and resuspending in 1 ml sterile 0.85% NaCl; repeat once.
- Resuspend pellet in 100 μl of sterile 0.85% NaCl.
- Individually spot 10 μl of washed culture for each strain on a plain TY agar plate. These spots are control spots.
- Add 40 μl of each strain excluding the E. coli DH5α12 containing pJC2 in a sterile tube, mix, and spot 120 μl of this mixture on a plain TY agar plate. This spot is the no-integrase negative control.
- Add 40 μl of each strain in a sterile tube, mix, and spot 160 μl of this mixture on a plain TY agar plate. This spot is the IMCE mating spot.
- Allow spots to dry in a laminar flow hood.
- Seal plates and incubate at 30 °C overnight.
3. Isolation of Trans-integrants
- Streak control spots and IMCE spot onto TY agar plates supplemented with 200 μg/ml streptomycin (SmUW227 is based on Rm1021 which carries a mutation conferring high-level resistance to streptomycin14) and 30 μg/ml gentamicin.
- For calculation of the IMCE efficiency, resuspend approximately ¼ of the IMCE mating spot in 500 μl sterile 0.85% NaCl. Make 10-fold serial dilutions to 10-7. Plate dilutions 10-2 through 10-5 on TY agar plates supplemented with 200 μg/ml streptomycin and 30 μg/ml gentamicin, to select for trans-integrants. Plate dilutions 10-3 through 10-7 on TY agar plates supplemented with 200 μg/ml streptomycin, to select for both trans-integrants and potential recipients, i.e. total recipients.
- For isolation of markerless trans-integrants resuspend approximately a quarter of the IMCE mating spot in 500 μl of sterile 0.85% NaCl. Make 10-fold serial dilutions to 10-6. Plate dilutions 10-4 to 10-6 on four TY agar plates supplemented with 200 μg/ml streptomycin and 200 μg/ml X-gluc.
- Incubate plates at 30 °C for 3 days.
- View RFP trans-integrants under green light (525 nm) and through a red filter (>610 nm) 15.
- Calculate the percent IMCE efficiency as CFU of trans-integrants compared to the CFU of total recipients. Approximately half of trans-integrants will have undergone true cassette exchange making them white and spectinomycin sensitive.
- To find the markerless trans-integrants (where the donor vector does not contain Gmr like in pJH110) screen for a white colony (an extended incubation period, 1-2 extra days at room temperature, will enhance the differentiation of colonies via X-gluc, this is necessary in S. meliloti SmUW227), confirm the colony's spectinomycin sensitivity by screening on an TY agar plate without antibiotics and an TY agar plate containing 100 μg/ml spectinomycin.
4. Representative Results
After three days of incubation on TY supplemented with streptomycin and gentamicin the control streaks should have no growth. The IMCE streak should have confluent growth on the head streak and many colonies on the second streak, as seen in Figure 2. The efficiency of trans-integration, expressed as the percentage of trans-integrants to total recipients, should be in the range of 0.5%. Approximately half of trans-integrants will be spectinomycin sensitive and white showing they have undergone true cassette exchange. Trans-integrants containing the tester rfp donor cassette from pJH110 should display discernable RFP fluorescence when viewed under green light (525 nm) and through a red filter (>610 nm) 15.
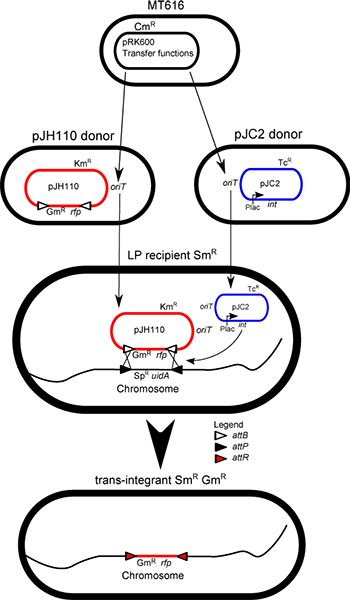
Figure 1. Conjugation mixture illustration: Aided by the expression of the transfer genes from pRK600, all of the plasmids are transferred randomly from cell to cell. This transfer results in the creation of trans-integrants in the mixture, through the LP-strain's acquisition of the two plasmids required for IMCE, the integrase (int) helper plasmid pJC2 and the donor plasmid pJH110. The donor cassette from the non-replicating donor plasmid (pJH110) is exchanged via ΦC31 integrase activity with the markers of the LP-cassette on the chromosome, resulting in the loss of the LP-markers (Spr and uidA) and the maintenance of the donor cassette (rfp and Gmr) in the resulting trans-integrant.
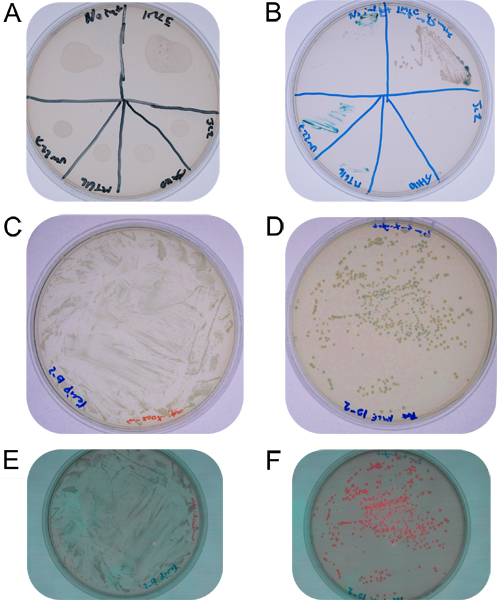
Figure 2. A: Mating spot plate on a non-selective TY plate showing dried cell mixtures on agar surface. B: Mating spots streaked on streptomycin-gentamicin-X-gluc plate, from top left clockwise no integrase control, IMCE into S. meliloti, E. coli DH5α containing pJC2 control, E. coli DH5a containing pJH110 control, E. coli MT616 control, and S. meliloti UW227 control. C: 10-2 dilution of mating spot resuspension on TY streptomycin-X-gluc agar. D: 10-2 dilution of mating spot resuspension on TY streptomycin-gentamicin-X-gluc agar (blue colonies are single recombinants, white colonies have undergone true cassette exchange). E: 10-2 dilution of mating spot resuspension on TY streptomycin-X-gluc agar showing lack of fluorescence. F: 10-2 dilution of mating spot resuspension on TY streptomycin-gentamicin-X-gluc agar showing two levels of fluorescence (brighter colonies correspond to blue colonies and have higher rfp expression presumably due to promoter read-though from vector sequence, where colonies having undergone true-cassette exchange contain RFP with only its immediate promoter with no read-through from the lac promoter in the vector, which is absent.).
Discussion
The IMCE technique allows for the efficient integration of a single attB flanked DNA cassette into the LP-locus of a previously engineered strain. Once the desired construct is cloned in place of rfp to create the donor cassette, the technique does not require subsequent DNA purification and transformation, making it very robust. It is critical that appropriate growth controls are included, to be certain the antibiotic resistance is due to the creation of trans-integrants and not other factors.
Disclosures
No conflicts of interest declared.
Acknowledgements
To Margaret C.M. Smith for kindly providing the integrase clone
Funding support from:
Genome Canada/Genome Prairie
NSERC Discovery and Strategic Project grants
Materials
| Name | Company | Catalog Number | Comments |
| Streptomycin | BioShop Canada | STP101 | |
| Spectinomycin | BioShop Canada | SPE201 | |
| Gentamicin | BioShop Canada | GTA202 | |
| Choramphenicol | BioShop Canada | CLR201 | |
| Tetracycline | BioShop Canada | TET701 | |
| Kanamycin | BioShop Canada | KAN201 | |
| Bacteriological grade agar | BioShop Canada | AGR001 | |
| Tryptone | BioShop Canada | TRP402 | |
| Yeast Extract | BioShop Canada | YEX401 | |
| Sodium Chloride | BioShop Canada | SOD001 | |
| Calcium Chloride | BioShop Canada | CCL444 | |
| X-gluc | Gold Biotechnology | G1281C1 | |
| E. coli MT616 strain | Available upon request | Also used outside of our lab | |
| E. coli pJC2 strain | In House | ||
| E. coli pJH110 strain | In House | ||
| SmUW227 strain | In House |
References
- Itaya, M., Tsuge, K., Koizumi, M., Fujita, K. Combining two genomes in one cell: Stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc. Natl. Acad. Sci. U.S.A. 102, 15971-15976 (2005).
- Kushtoss, S., Rao, R. N. Analysis of the Integration Function of the Streptomycete Bacteriophage FC31. J. Mol. Biol. 222, 897-908 (1991).
- Brown, W. R., Lee, N. C., Xu, Z., Smith, M. C. Serine recombinases as tools for genome engineering. Methods. 53, 372-379 (2011).
- Groth, A. C., Olivares, E. C., Thyagarajan, B., Calos, M. P. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. U.S.A. 97, 5995-6000 (2000).
- Rowley, P. A., Smith, M. C., Younger, E. A motif in the C-terminal domain of FC31 integrase controls the directionality of recombination. Nuc. Acid. Res. 36, 3879-3891 (2008).
- Campbell, R. E. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 99, 7877-7882 (2002).
- Schafer, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutumicum. Gene. 145, 69-73 (1994).
- Thorpe, H. M., Smith, M. C. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. 95, 5505-5510 (1998).
- Jones, J. D., Gutterson, N. An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens strain HV37a. Gene. 61, 299-306 (1987).
- Beringer, J. E. R Factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84, 188-198 (1974).
- Lennox, E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1, 190-206 (1955).
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557-580 (1983).
- Charles, T. C., Finan, T. M. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J. Bacteriol. 172, 2469-2476 (1990).
- Leigh, J. A., Signer, E. R., Walker, G. C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U.S.A. 82, 6231-6235 (1985).
- Heil, J. R., Nordeste, R. F., Charles, T. C. The fluorescence theatre: a cost-effective device using theatre gels for fluorescent protein and dye screening. Can. J. Microbiol. 57, 339-342 (2011).
- Datsenko, K. A., Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640-6645 (2000).
- Lesic, B., Rahme, L. G. Use of the lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol. Biol.. 9, 20-20 (2008).
- Choi, K. -. H., Schweizer, H. P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protocols. 1, 153-161 (2006).
- Thomason, L. C., Calendar, R., Ow, D. W. Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage FC31 site-specific recombination system. Molecular Genetics and Genomics. 265, 1031-1038 (2001).
- Katzen, F. Gateway recombinational cloning: a biological operating system. Expert Opin. Drug Discovery. , 571-586 (2007).
- Charles, T. C., Doty, S. L., Nester, E. W. Construction of Agrobacterium strains by electroporation of genomic DNA and its utility in analysis of chromosomal virulence mutations. Appl. Environ. Microbiol. 60, 4192-4194 (1994).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved