A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Process Optimization using High Throughput Automated Micro-Bioreactors in Chinese Hamster Ovary Cell Cultivation
In This Article
Summary
Here, we present a detailed procedure to run a Design of Experiment in an automated micro-bioreactor followed by cell harvest and protein quantification using a Protein A column.
Abstract
Optimization of bioprocesses to increase the yield of desired products is of importance in the biopharmaceutical industry. This can be achieved by strain selection and by developing bioprocess parameters. Shake flasks have been used for this purpose. They, however, lack the capability to control the process parameters such as pH and dissolved oxygen (DO). This limitation can be overcome with the help of an automated micro-bioreactor. These bioreactors mimic cultivation at a larger scale. One of the major advantages of this system is the integration of the Design of Experiment (DOE) in the software. This integration enables establishing a design where multiple process parameters can be varied simultaneously. The critical process parameters and optimum bioprocess conditions can be analyzed within the software. The focus of the work presented here is to introduce the user to the steps involved in process design in the software and incorporation of the DOE within the cultivation run.
Introduction
The global biopharmaceutical market was worth more than US $250 billion in 2018 and has been continuously expanding1. Pharmaceutical companies are moving away from producing small molecular drugs to biotechnologically produced therapeutics such as recombinant proteins. These alone are responsible for a revenue of more than $150 billion1. Mammalian cells are now extensively used for the production of these pharmaceutical recombinant proteins. In the current period, among the 68 approved products produced by mammalian cells, 57 are produced by Chinese Hamster ovary cells (CHO)2. CHO cells are specifically used for the production of recombinant proteins that require post-translational modifications. These cells are preferred as they grow in a suspension and thereby enable reproducible results in a serum free chemically defined medium3,4. The other advantage of using CHO cells is that the glycan structure of the product resembles that of the human monoclonal antibody (mAb) and results in higher recombinant protein yield and specific productivity due to gene amplification5.
The yield of recombinant CHO (rCHO) cell culture has increased by a hundred-fold in the past two decades. This improvement is attributed to the optimization of the process parameters, feeding strategy and development of serum free chemically defined medium6. With the increase in requirements of the pharmaceutical products, the pressure increases on cost and time efficiency for the development of the production process7. To reduce the pressure while assuring product quality has redirected the focus of the pharmaceutical industry on Quality by Design (QbD). QbD is used to understand the product production as well as the process. A vital tool used in the ObD is the Design of Experiment (DOE). It helps increase understanding of the process by revealing the relationship between various input variables and resulting output data. Applying the DOE approach to optimize the bioprocess is beneficial during the early stages of the project in assimilating the process conditions and increasing the titer quantity and quality. This approach is beneficial when compared to the old-fashioned strategy: one-factor-at-a-time (OFAT). The statistical approaches to DOE using Classical, Shainin or Taguchi are far superior to the OFAT8.
The process and media optimization can be performed in shake flasks. The flasks are relatively inexpensive. However, it is not possible to control parameters such as temperature, pH and dissolved oxygen (DO). To overcome these drawbacks, multiuse bench-top bioreactors ranging from working volume of 0.5 L to 5 L can be used. The reactors provide an extensive on-line monitoring and process control. However, the use of the multiuse bioreactor is time and labor intensive. In order to overcome these disadvantages, a novel single-use bioreactor that combines the comprehensive process of monitoring the bench-top bioreactor and easy handling of the shake flask is used. The high throughput screening system and single-use technology have contributed to enhance the efficiency of process performance and development9.
In this article, the guidelines to load the recipe in the automated micro-bioreactor (AMBR) software are listed. The influence of different stirrer speeds and pH on the viable cell concentration (VCC) and titer is studied during the course of this experiment. The experimental result and analysis are carried out with design of experiment software MODDE 12. The product analytics are carried out in a high pressure liquid chromatography (HPLC) system with a Protein A column. It is based on the principle that the Fc region of the mAb binds to protein A with high affinity10,11. With this method, it is possible to identify and quantify the mAb. The quantification is carried out over the measured elution peak areas at 280 nm.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Preculture Procedure
NOTE: Recombinant CHO DG44 cells with a viable cell concentration of 1 x 107 cells/mL are used for this protocol.
- Thaw the vial containing 1.2 mL of cells to room temperature and immediately transfer the cell suspension to a 15 mL conical centrifuge tube containing 10 mL of cold seed medium.
- Centrifuge the conical centrifuge tube for 5 minutes at 190 x g and room temperature and discard the supernatant.
- Pre-heat 150 mL of the seed medium in a 500 mL shake flask to 36.8 °C.
- Gently resuspend the cell pellet in 10 mL of pre-warmed seed medium and transfer the cells into the shake flask.
- Use 1 mL of the sample from the flask to measure the initial VCC and viability using a cell counter.
NOTE: The viability should be above 70% after thawing for successful cultivation. - Incubate the shake flask in an orbital shaker (shaker diameter of 19 mm) at 36.8 °C and 7.5% CO2 with a shaking rate of 120 rpm.
NOTE: These conditions vary depending on the cell strain and medium. - Three days after passaging the cells, remove the shake flask from the shaker and place it under the laminar flow cabinet. Take 1 mL of sample to measure the final cell concentration. Calculate the volume to be transferred to fresh pre-warmed seed medium such that the initial cell concentration in the new passage is 2 x 105 cells/mL.
- Passage the cells 5 times in total before transferring to the bioreactor for the main cultivation.
2. Main Cultivation
- Measure the final cell concentration of the preculture. Calculate the volume to be transferred to the bioreactor such that the initial cell concentration in the reactor is 3 x 105 cells/mL.
- Fill the reactor with production medium a day before the inoculation to equilibrate the reactor and set the process parameters such as temperature, pH and DO.
NOTE: The cultivation conditions are 36.8 °C and 60% dissolved oxygen concentration (DO). We tested stirrer speeds of 1050 rpm and 1300 rpm along with pHs of 6.9, 7.1, and 7.3. The total duration of the cultivation is 12 days until the cells are harvested. The batch process runs for 72 hours after which the feed medium is added every 24 h. The protocol to be used for the cultivation is listed in detail in the next segment.
3. Writing the Recipe in the Automated Micro-bioreactor Software
NOTE: There are two ways of writing a recipe in the AMBR cell culture software: it is created either by using a wizard or by adding each step manually. For the purpose of this protocol, steps using the wizard are shown.
- Creating a new experiment
- Open the AMBR cell culture software and in the Introduction tab click on Create New Experiment.
- Loading the recipe
- In the New Experiment tab, enter the name of the experiment along with the date on which it is to be conducted.
- Activate the check point for the culture station and the vessels to be used during the cultivation. The Auto Add DOE Tags will also be activated for an easy transition during the programming of the DOE experiment. Click on Next to switch to the next tab.
- Set information about addition of media into the vessel along with antifoam, inoculum, feed and glucose.
- Activate the Add Media Plate check point. Define the Plate type, name and location of the plate containing the medium.
CAUTION: Depending on the type of plate and if the plate contains a lid, activate the check on Is Lidded to ensure smooth functioning of the liquid handler - Click on Add Media To Vessels. Enter the volume of the media to be added into the vessels. Define the mapping of the transfer of the media from the plate to the vessels. Click on Next to switch to the next tab.
- Activate the Add Media Plate check point. Define the Plate type, name and location of the plate containing the medium.
- Set the cultivation conditions in the reactor.
- After the media information has been fed into the software, assign the cultivation conditions. Click on Condition Media and fill in the temperature, target DO, upper pH limit and stirring RPM (Up stirring or Down stirring).
- Set addition of inoculums into the vessels.
- Activate Add Cell Plate. Define the plate type, name and location of the plate containing the medium.
- Click on Add Cells To Vessels. Enter the time of inoculation and the volume of the media to be added to the vessels.
- Define the path travelled by the liquid handler to the transfer of the cell from the plate to the vessels. Click on Next to switch to the next tab.
NOTE: Ensure Reuse Pipette Tips is deactivated to avoid cross-contamination and incorrect initial viable cell concentration.
- Set addition of feed, glucose and antifoam.
NOTE: The procedure for addition of feed, glucose and antifoam is similar to each other. For the sake of this protocol the procedure is listed for “Feed”. This can be replicated for glucose and antifoam.- Activate the Add Feed Plate and define the plate type, name and location. Click on Add Feed To Vessels and enter the volume of the feed to be added to the vessels. Define the mapping of the transfer of the feed from the plate to the vessels.
- Depending on the cultivation, add the number of feed addition. For this cultivation, the reactor is fed after 72 hours for every 24 hours.
- Manually add the time delay between the feeding by entering the data into Delay from cells added. The first day of feeding is after 72 hours of inoculation and the next one is after 96 hours and so on.
NOTE: Antifoam addition is programmed to be added every day to avoid foaming during the cultivation.
- Set sampling during the cultivation.
- Activate the Add Sample Plate and define the plate type, name and location.
- Check on Take Sample from Vessels and enter the volume of the sample to be removed from the vessels. Define the mapping of the transfer of the sample from the vessels to the plate. Ensure that the volume does not decrease below 10 mL during the entire course of cultivation.
- Add the number of samples to be taken during the cultivation. Similar to feeding, add the time of the sample being removed from the vessel for each input sample point.
- Save the process. It is now ready for execution.
NOTE: To ensure the smooth running of the protocol, switch to the Process Steps tab in the AMBR cell culture software and select Process Step view to visualize the flow of the recipe.
- In the New Experiment tab, enter the name of the experiment along with the date on which it is to be conducted.
- Design of experiment in the automated micro bioreactor
- In order to run the DOE software of the bioreactor, ensure that the recipe in the main software is saved and ready to use.
- Open the AMBR 15 DOE software and click on Investigation and select New.
- Enter the name of the new DOE investigation in the Create Investigation dialog box.
- In order to assign an experiment to the DOE investigation, open the recipe created to study the different parameters. Click on Browse and select the respective experiment.
- Define the DOE factor.
- The vessel tags are already enlisted in the column. To define the desired DOE factor, select the parameter and click on the column labeled DOE factor. Select New and add the units, abbreviation, lower and upper limit of the factors (e.g., temperature, DO, pH).
- Define the response factor.
- Once the DOE factors have been defined, define the response based on which the experimental analysis would be structured.
- In the Responses tab, define the values to be considered for the analysis of the data.
- Click on Edit DOE Responses and define the name of the response, abbreviation, units, minimum and maximum limits (e.g., titer, viable cell concentration).
- Once the responses are defined, select the AMBR variable for each response and define the variable. A response can be automatically associated with a micro-bioreactor variable, Choose the required variable from the drop-down list.
- Change the equation for each of the response depending on the requirement. The choice is between the minimum, maximum, first, last and average data.
- Create a design.
- Use the Start Design Wizard in order to select the type of experimental design, to add or remove the number of replicates and center points.
- Select the objective, which determines the choice of designs and models:
Screening: Uses linear and interaction models to find the important factors
Optimization (RSM), Uses quadratic and cubic models for detailed modeling and optimization
Split objective: Models for formulation and process factors can be chosen separately - Once the objective is decided upon, select the model and the design along with the number of center points and replicates.
- Click on Finish and switch to the next tab.
- Define the experiment.
NOTE: The DOE factors are listed in the right column of the software. On selecting the desired factors, the vessels running that experiment with the desired parameter would be highlighted. The vessels within the culture station can be moved around by right clicking on the vessel and moving it to the desired location.- Create work packets that can be imported in the AMBR cell culture software. Depending on the number of experiments the different work packets are created and stored for further implementation
- Execution of the experiment in the work packets created on the AMBR control laptop
- In the Experiment tab, click on Create DOE Experiment and browse for the work packet created using the DOE software.
- Initialize the process by clicking Start.
- Analysis of experimental results
- Once the experiment has been executed, export the data using Export DOE Results. The Export DOE Results window opens and the rows indicating the culture vessel and station are listed in the table.
- Select the desired rows and click on Exported Selected Rows or Export Experimental data to store all the results and save the file for further analysis.
- Import the data into the AMBR DOE module by switching to the Results tab and selecting Import Results.
- Browse for the desired data file and click the Analysis Results.
- Analyze the results further in MODDE.
4. Execution of Cultivation in the Automated Micro-bioreactor
NOTE: The following steps are executed by the user with the help of the protocol written in the aforementioned software. The steps are carried out by the user unless mentioned otherwise.
- Loading the vessels
- Open the gamma sterilized culture vessels under a laminar flow cabinet and orient in the culture station as depicted in Figure 2.
- Clean the clamp plate with 70% ethanol and double distilled water. Then, autoclave the plate and place on top of the vessel.
- Mount the clamp plate with a stirrer plate, ensuring each pin is fixed firmly.
- Tighten both the stirrer plate and clamp plate onto the stirring assembly.
- Running the micro-bioreactor software
- Use the program written in Section 3 to run the cultivation.
- Visualize the process steps scheduled or completed in the Process tab. Alter the cultivation steps during the process run as needed by first pausing the liquid handler and then editing the process recipe.
- Add antifoam to the vessels before the stirrer is started to ensure there is no excessive foaming during the cultivation. The antifoam will be added regularly, and the foam detected visually.
- Addition of media into the vessel
- Fill the 24 well plate provided with the micro-bioreactors manually with sterile media and place in the designated deck of the system. Ensure the plate is placed in the deck designated by the written program (section 3). The filling of the vessel will take place as designed in section 3.2.2.
NOTE: The temperature and stirrer start immediately after the addition of the media and the antifoam. The sensor reader is activated 1 hour after the vessel is filled (Start Monitor step). Gassing to each vessel commences once the reader has been activated. Media is left to equilibrate for a minimum of 6 hours before pH recalibration and inoculation. The process parameters can be altered in the software as mentioned in section 3.2.3.
- Fill the 24 well plate provided with the micro-bioreactors manually with sterile media and place in the designated deck of the system. Ensure the plate is placed in the deck designated by the written program (section 3). The filling of the vessel will take place as designed in section 3.2.2.
- Inoculation
- Measure the viable cell concentration after the 5th passage. Calculate the number of cells to be transferred to the vessels to ensure that the initial cell concentration in all the vessels is 3 x 105 cells/mL.
- Transfer the cells to a 24 deep well plate such that the volume of the suspension is at least 1.6 times the required volume. For a required volume of 2 mL of the inoculum, transfer 3.2 mL of cell suspension into each well in the plate.
- Place the 24 well plate in the designated deck. The vessels will be inoculated as in section 3.2.4.
- Daily sampling and analytics
- Remove a 460 μL sample from the vessels every day using the liquid handler. Dilute 200 µL of the sample with 800 µL of filtered 1x PBS buffer (5x dilution), and then place in the cell counter.
- Centrifuge the remaining sample for 5 min at 190 x g and room temperature and store the supernatant for further analysis (glucose, lactate, glutamine and glutamate).
- Freeze 100 µL of the supernatant at -20 °C until the end of the cultivation for protein quantification.
- End of cultivation
- When the process parameter control (i.e., temperature, agitation, pH and DO) has terminated, stop the monitoring of the process.
- Unscrew the stirrer plate and clamp plate.
- Remove the culture vessels and clean the culture stations. Place the drying plates on the culture stations and screw them in.
- Meanwhile, clean the clamp plates thoroughly with 70% ethanol and double distilled water.
- Click on Stop in the bioreactor software once the drying cycle is completed.
- Cell culture harvest
- Harvest cells on Day 12 of the cultivation by manually removing the content of the vessels into 50 mL centrifuge tubes. Centrifuge the cell broth at 190 x g for 30 min.
- Discard the cell pellet and store the supernatant at -20 °C.
5. Measuring mAb Concentration
- Use a 1.7 mL Protein A column for the quantification of the protein during the cultivation run.
- Prepare the equilibration and elution buffer before thawing the samples.
- Use a solution of 0.5 M Na2HPO4 containing 0.5 M NaCl with a pH of 7.9 as the equilibration buffer and a solution of 100 mM glycine containing 0.5 M NaCl with a pH of 2 as the elution buffer.
- Filter both buffers through a 0.2 µm membrane and degas before being placed for the analysis.
- Purge the high-performance liquid chromatography system (HPLC) with freshly prepared equilibration buffer.
- Load the protein A column on to the HPLC system.
- Carry out chromatography with a flow rate of 1 mL/min. Set the column oven temperature at 30 °C and autosampler temperature at 10 °C
- Thaw the frozen samples at room temperature and filter 225 µL of each sample through a 0.22 µm PVDF membrane. Dilute the samples with higher concentration of the desired protein are in a 1:20 ratio with equilibration buffer and filter through the membrane before placing in the autosampler.
- Place the samples in the autosampler. Load the method and sequence in the software and start the sequence.
NOTE: The method is comprised of three phases (see Figure 1): injection of the sample into the column for the first two minutes; followed by elution buffer for 8 min and column regeneration with equilibration buffer for 10 min.
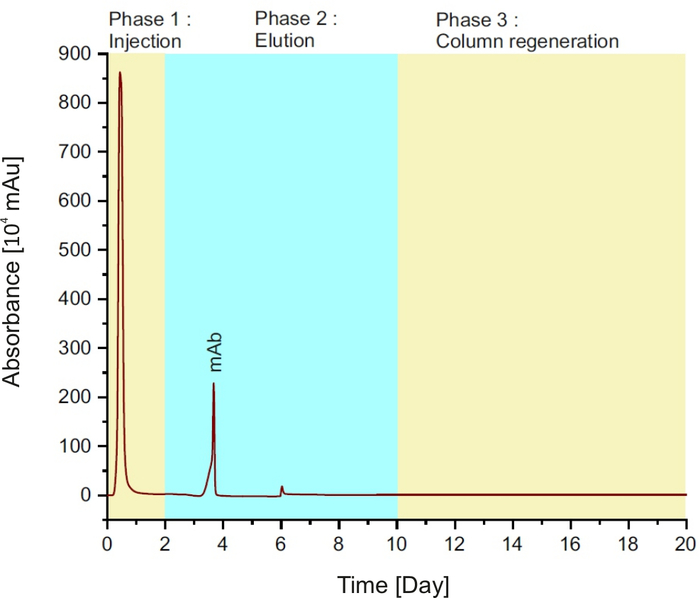
Figure 1: Protein A chromatogram, representing the different phases during a single run. Please click here to view a larger version of this figure.
Access restricted. Please log in or start a trial to view this content.
Results
An overview of the cultivation performed in this study is presented in Figure 2.
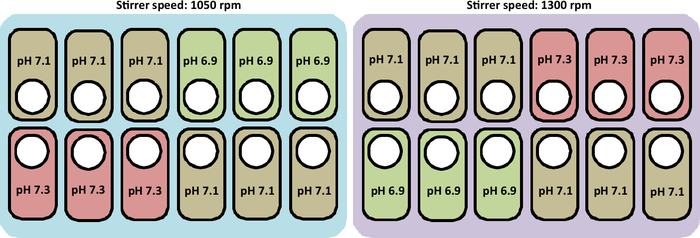
Figure 2: Schematic representation of the experimental conditions to test pH and stirrer speed profiles in the culture stations. The figure also represents the correct layout to place the vessels.
Access restricted. Please log in or start a trial to view this content.
Discussion
Optimization of the process to increase the yield is of crucial importance in the biopharmaceutical industry. Shake flasks could potentially be used for the screening of the strain; however, the monitoring of the process parameters such as pH and DO are unavailable in the flasks. The micro-bioreactors have an advantage as they allow continuous monitoring and control of the process. These control loops in the micro-bioreactor also provide a condition similar to those at larger scale and thus, deliver results that are comp...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank the Bundesministerium für Bildung und Forschung (BMBF), the Federal Ministry of Education and Research, Germany, and the BioProcessing team of Sartorius Stedim Biotech GmbH, Germany, for their support.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 1 mL disposable pipette tips, sterilized | Sartorius Stedim Biotech GmbH | A-0040 | |
| 200 mM L-glutamine | Corning, Merck | 25-005-CV | |
| 24 Well deep well plates | Sartorius Stedim Biotech GmbH | A-0038 | |
| 5 mL disposable pipette tips, sterilized | Sartorius Stedim Biotech GmbH | A-0039 | |
| ambr 15 automated microbioreactor system | Sartorius Stedim Biotech GmbH | 001-2804 | |
| ambr 15 Cell Culture 24 Disposable Bioreactors - Sparged | Sartorius Stedim Biotech GmbH | 001-1B86 | |
| Antifoam C Emulsion | Sigma-Aldrich, Merck | A8011 | |
| Bottle Top Sterile filter | Corning, Merck | CLS431474 | 0.1 μm pore size |
| CEDEX Detergent (3% Mucosol) | Roche Innovatis AG | 05-650-658-001 | |
| Cell counter | Roche Innovatis AG | 05-650-216-001 | CEDEX HiRes |
| CHO DG44 cell line | Cellca, Sartorius Stedim Biotech GmbH | ||
| CHOKO Feed Media A (FMA) | Sigma-Aldrich, Merck | CR80025 | |
| CHOKO Feed Media B (FMB) | Sigma-Aldrich, Merck | CR80026 | |
| CHOKO Production Medium | Sigma-Aldrich, Merck | CR80027 | |
| CHOKO Stock Culture Meium | Sigma-Aldrich, Merck | CR80028 | |
| Chromaster high pressure liquid chromatography system | VWR International | ||
| Conical Centrifuge tube | Corning, Merck | SIAL0790 | |
| Ethanol | Merck | 1070179026 | |
| Glycine | Carl Roth | 56-40-6 | |
| HPLC Vials | VWR International | SUPLSU860181 | |
| PBS | Sigma-Aldrich,Merck | P4417 | |
| Protein A Column | Thermo Fisher Scientific | 1502226 | POROS™ A 1.7 mL |
| Sodium chloride | Sigma-Aldrich,Merck | 7647-14-5 | |
| Sodium phosphate dibasic anhydrous | Sigma-Aldrich,Merck | 7558-79-4 | |
| Trypan Blue | VWR International | VWRVK940 | |
| YSI | YSI Inc | 2900D | YSI 2900 Select |
References
- Langer, E. S. 15th Annual report and survey of Biopharmaceutical Manufacturing Capacity and Production: A study of Biotherapeutical Developers and Contract Manufacturing Organizations. Bioplan Associates. , Available from: http://bioplanassociates.com/wp-content/uploads/2018/07/15thAnnualBiomfgReport_TABLEOFCONTENTS-LR.pdf (2019).
- Walsh, G. Biopharmaceutical benchmarks 2018. Nature Biotechnology. 36, 1136(2018).
- Kim, J. Y., Kim, Y., Lee, G. M. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Applied Microbiology and Biotechnology. 93 (3), 917-930 (2012).
- Lai, T., Yang, Y., Ng, S. K. Advances in Mammalian cell line development technologies for recombinant protein production. Pharmaceuticals (Basel). 6 (5), 579-603 (2013).
- Carlage, T., et al. Analysis of dynamic changes in the proteome of a Bcl-XL overexpressing Chinese hamster ovary cell culture during exponential and stationary phases. Biotechnology Progress. 28 (3), 814-823 (2012).
- Hacker, D. L., de Jesus, M., Wurm, F. M. 25 years of recombinant proteins from reactor-grown cells - where do we go from here. Biotechnology Advances. 27 (6), 1023-1027 (2009).
- Shukla, A. A., Gottschalk, U. Single-use disposable technologies for biopharmaceutical manufacturing. Trends in Biotechnology. 31 (3), 147-154 (2013).
- Ao, S., Gelman, L. Advances in electrical engineering and computational science. Lecture notes in electrical engineering. 39, Springer. New York. (2009).
- Bareither, R., et al. Automated disposable small scale reactor for high throughput bioprocess development: a proof of concept study. Biotechnology and Bioengineering. 110 (12), 3126-3138 (2013).
- Kang, Y., Ludwig, D. L., Balderes, P. What can cell culture flocculation offer for antibody purification processes. Pharmaceutical Bioprocessing. 2 (6), 483-485 (2014).
- Choe, W., Durgannavar, T. A., Chung, S. J. Fc-Binding Ligands of Immunoglobulin G: An Overview of High Affinity Proteins and Peptides. Materials (Basel). 9 (12), (2016).
- Schäpper, D., et al. Application of microbioreactors in fermentation process development: a review. Analytical and Bioanalytical Chemistry. 395 (3), 679-695 (2009).
- Zhang, Z., et al. Microbioreactors for Bioprocess Development. Journal of the Association for Laboratory Automation. 12 (3), 143-151 (2007).
- Claßen, J., et al. Spectroscopic sensors for in-line bioprocess monitoring in research and pharmaceutical industrial application. Analytical and Bioanalytical Chemistry. 409 (3), 651-666 (2017).
- Janoschek, S., et al. A protocol to transfer a fed-batch platform process into semi-perfusion mode: The benefit of automated small-scale bioreactors compared to shake flasks as scale-down model. Biotechnology Progress. 35 (2), 2757(2019).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved